Gene
KWMTBOMO12816
Pre Gene Modal
BGIBMGA012742
Annotation
PREDICTED:_protein_vein_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.547 Nuclear Reliability : 1.679
Sequence
CDS
ATGTTTATTCTCACAGGTATTCCAGCCCGAGTGGAGCTTATGCCGTCAGTGTTGACTACCAAGGGCGGGAGGGTGCGACTGGAGTGTAAAGCCAGCGCGCGACCTCCGCCGCGCATCACGTGGTACAAGAACGGCAAGCCTGTCGCCGATATCGCTTTGAGGAGACTCCGGGTGCAAAATTACAGAAGGCGCTCGGTATTAGTGATAAGGCACGCAAAACTAAACGATACAGCGAGCTATGAATGCAAGGCCCAAGGTGCTATTGGTCCTCCTGCCATCGCGACGGCCAACGTCACCGTCCTCGCGCCTCCATCAGATCCGACGTCTGGGTCACAGTGTCCAATGCCGGACCCTTCCTCTTACTGCCTCAACGGAGGCACTTGCATATTTTTTGAAATGCTGCGAGAGCAGGCATGCACATGTAAAGATGGATTCGGGGGGCAGCGATGCGAGATCAAGGACGTTAAAAGCAGGAGTAGCAAAGGGGAGCAGCTTTGGTAG
Protein
MFILTGIPARVELMPSVLTTKGGRVRLECKASARPPPRITWYKNGKPVADIALRRLRVQNYRRRSVLVIRHAKLNDTASYECKAQGAIGPPAIATANVTVLAPPSDPTSGSQCPMPDPSSYCLNGGTCIFFEMLREQACTCKDGFGGQRCEIKDVKSRSSKGEQLW
Summary
Uniprot
H9JT80
A0A3S2NE06
A0A2W1BX89
A0A2A4JNB5
A0A2H1VE17
A0A212F2X8
+ More
A0A194RG63 A0A194QDW1 I4DS55 A0A146LTY7 W5JG68 A0A0A9Z4I1 A0A182FGX4 A0A1B0EW48 A0A1L8E3H7 A0A2S2QVR8 A0A2H8TJJ8 A0A0L7RIJ2 J9L3Y9 A0A2S2P1B5 A0A2H8TZW1 A0A147BAL4 A0A1I8NWL6 A0A0P5KJM5 A0A0N8CGS5 A0A0N8DG88 A0A2H8TCQ5 A0A0P5TCN8 A0A1B6LM22 A0A0P5J843 A0A1Z5KY15 A0A0P5C6R8 A0A0N8BJI2 A0A0P6GEZ0 A0A0P5JDV0 A0A0P5JFU2 A0A0P5D4V7 A0A0P6FL89 A0A0P5L055 A0A0P5K3C2 A0A0P5WTG3 A0A0P5WYN8 A0A0P5NZA8 A0A0P4YQE0 A0A0N8BSL2 A0A1B6IHR8 A0A0P5Q670 A0A0P5CJD5 A0A0P5RIX1 A0A0P5JM69 A0A0P5IZU0 A0A0N8BWF6 A0A0P6F9D2 A0A1B6F5Q1 A0A0P6INH2 A0A0N7ZUU5 A0A0P6BNH8 A0A0P5N3U6 A0A0P5NM38 A0A034WFV6 A0A1A9XR29 A0A1B0ALJ4 A0A182MSU3 A0A1A9Z950 A0A1A9W4W3 T1KWH2 A0A182WLH1 A0A0N7ZLM7 A0A088AHR5 A0A1S3D240 A0A1J1I2R7 A0A0L0BWA0 A0A182NK73 A0A0A1WDH0 A0A0P4ZSF2 A0A1B0DNQ4 A0A443RED3 A0A1B6DH23 A0A1B0FM61 A0A182SRU4 A0A1W4WPE5 A0A1W4WPF3 A0A084VZ66 A0A3B0JUW6 A0A1W4X074 A0A2C9H5T1 A0A2C9GPT8 A0A1W4X0F6 Q29E55 B4HAV9 A0A0N7ZNM1 A0A1S4GUY5 A0NBJ8 A0A1W4WZ61 A0A3Q2CZ96 B7PKT5 A0A3B3RBE4 A0A330IZ60
A0A194RG63 A0A194QDW1 I4DS55 A0A146LTY7 W5JG68 A0A0A9Z4I1 A0A182FGX4 A0A1B0EW48 A0A1L8E3H7 A0A2S2QVR8 A0A2H8TJJ8 A0A0L7RIJ2 J9L3Y9 A0A2S2P1B5 A0A2H8TZW1 A0A147BAL4 A0A1I8NWL6 A0A0P5KJM5 A0A0N8CGS5 A0A0N8DG88 A0A2H8TCQ5 A0A0P5TCN8 A0A1B6LM22 A0A0P5J843 A0A1Z5KY15 A0A0P5C6R8 A0A0N8BJI2 A0A0P6GEZ0 A0A0P5JDV0 A0A0P5JFU2 A0A0P5D4V7 A0A0P6FL89 A0A0P5L055 A0A0P5K3C2 A0A0P5WTG3 A0A0P5WYN8 A0A0P5NZA8 A0A0P4YQE0 A0A0N8BSL2 A0A1B6IHR8 A0A0P5Q670 A0A0P5CJD5 A0A0P5RIX1 A0A0P5JM69 A0A0P5IZU0 A0A0N8BWF6 A0A0P6F9D2 A0A1B6F5Q1 A0A0P6INH2 A0A0N7ZUU5 A0A0P6BNH8 A0A0P5N3U6 A0A0P5NM38 A0A034WFV6 A0A1A9XR29 A0A1B0ALJ4 A0A182MSU3 A0A1A9Z950 A0A1A9W4W3 T1KWH2 A0A182WLH1 A0A0N7ZLM7 A0A088AHR5 A0A1S3D240 A0A1J1I2R7 A0A0L0BWA0 A0A182NK73 A0A0A1WDH0 A0A0P4ZSF2 A0A1B0DNQ4 A0A443RED3 A0A1B6DH23 A0A1B0FM61 A0A182SRU4 A0A1W4WPE5 A0A1W4WPF3 A0A084VZ66 A0A3B0JUW6 A0A1W4X074 A0A2C9H5T1 A0A2C9GPT8 A0A1W4X0F6 Q29E55 B4HAV9 A0A0N7ZNM1 A0A1S4GUY5 A0NBJ8 A0A1W4WZ61 A0A3Q2CZ96 B7PKT5 A0A3B3RBE4 A0A330IZ60
Pubmed
EMBL
BABH01034788
BABH01034789
RSAL01000176
RVE45041.1
KZ149907
PZC78265.1
+ More
NWSH01000922 PCG73527.1 ODYU01002052 SOQ39083.1 AGBW02010663 OWR48090.1 KQ460416 KPJ14926.1 KQ459324 KPJ01651.1 AK405427 BAM20745.1 GDHC01008417 JAQ10212.1 ADMH02001610 ETN61809.1 GBHO01006894 JAG36710.1 AJWK01005322 AJWK01005323 AJWK01005324 GFDF01000952 JAV13132.1 GGMS01012653 MBY81856.1 GFXV01002395 MBW14200.1 KQ414583 KOC70685.1 ABLF02040729 GGMR01010097 MBY22716.1 GFXV01007237 MBW19042.1 GEGO01007603 JAR87801.1 GDIQ01188394 JAK63331.1 GDIP01133247 JAL70467.1 GDIP01036743 JAM66972.1 GFXV01000092 MBW11897.1 GDIP01131993 JAL71721.1 GEBQ01015196 JAT24781.1 GDIQ01205256 JAK46469.1 GFJQ02006991 JAV99978.1 GDIP01174683 JAJ48719.1 GDIQ01171603 JAK80122.1 GDIQ01036905 JAN57832.1 GDIQ01203556 JAK48169.1 GDIQ01225333 JAK26392.1 GDIP01161224 JAJ62178.1 GDIQ01046607 JAN48130.1 GDIQ01202402 JAK49323.1 GDIQ01195242 JAK56483.1 GDIP01082043 JAM21672.1 GDIP01079688 JAM24027.1 GDIQ01156962 JAK94763.1 GDIP01225420 JAI97981.1 GDIQ01148963 JAL02763.1 GECU01025871 GECU01021234 GECU01000086 JAS81835.1 JAS86472.1 JAT07621.1 GDIQ01125426 JAL26300.1 GDIP01174684 JAJ48718.1 GDIQ01201097 GDIQ01100136 JAL51590.1 GDIQ01201096 GDIQ01100135 JAK50629.1 GDIQ01231586 JAK20139.1 GDIQ01138211 JAL13515.1 GDIQ01164171 GDIQ01063693 JAN31044.1 GECZ01024262 JAS45507.1 GDIQ01002825 JAN91912.1 GDIP01210377 JAJ13025.1 GDIP01012474 JAM91241.1 GDIQ01147423 JAL04303.1 GDIQ01147422 JAL04304.1 GAKP01006304 JAC52648.1 JXJN01031202 AXCM01003036 CAEY01000644 GDIP01233321 JAI90080.1 CVRI01000035 CRK92665.1 JRES01001347 KNC23509.1 GBXI01017248 JAC97043.1 GDIP01212380 JAJ11022.1 AJVK01017655 AJVK01017656 AJVK01017657 AJVK01017658 AJVK01017659 NCKU01000934 RWS13620.1 GEDC01012383 JAS24915.1 CCAG010015251 CCAG010015252 CCAG010015253 CCAG010015254 ATLV01018612 ATLV01018613 KE525239 KFB43260.1 OUUW01000002 SPP77499.1 APCN01000156 APCN01000157 CH379070 EAL30207.3 CH479244 EDW37741.1 GDIP01227769 JAI95632.1 AAAB01008807 EAU77664.2 ABJB010764675 DS735578 EEC07207.1 LT716783 SIW59160.1
NWSH01000922 PCG73527.1 ODYU01002052 SOQ39083.1 AGBW02010663 OWR48090.1 KQ460416 KPJ14926.1 KQ459324 KPJ01651.1 AK405427 BAM20745.1 GDHC01008417 JAQ10212.1 ADMH02001610 ETN61809.1 GBHO01006894 JAG36710.1 AJWK01005322 AJWK01005323 AJWK01005324 GFDF01000952 JAV13132.1 GGMS01012653 MBY81856.1 GFXV01002395 MBW14200.1 KQ414583 KOC70685.1 ABLF02040729 GGMR01010097 MBY22716.1 GFXV01007237 MBW19042.1 GEGO01007603 JAR87801.1 GDIQ01188394 JAK63331.1 GDIP01133247 JAL70467.1 GDIP01036743 JAM66972.1 GFXV01000092 MBW11897.1 GDIP01131993 JAL71721.1 GEBQ01015196 JAT24781.1 GDIQ01205256 JAK46469.1 GFJQ02006991 JAV99978.1 GDIP01174683 JAJ48719.1 GDIQ01171603 JAK80122.1 GDIQ01036905 JAN57832.1 GDIQ01203556 JAK48169.1 GDIQ01225333 JAK26392.1 GDIP01161224 JAJ62178.1 GDIQ01046607 JAN48130.1 GDIQ01202402 JAK49323.1 GDIQ01195242 JAK56483.1 GDIP01082043 JAM21672.1 GDIP01079688 JAM24027.1 GDIQ01156962 JAK94763.1 GDIP01225420 JAI97981.1 GDIQ01148963 JAL02763.1 GECU01025871 GECU01021234 GECU01000086 JAS81835.1 JAS86472.1 JAT07621.1 GDIQ01125426 JAL26300.1 GDIP01174684 JAJ48718.1 GDIQ01201097 GDIQ01100136 JAL51590.1 GDIQ01201096 GDIQ01100135 JAK50629.1 GDIQ01231586 JAK20139.1 GDIQ01138211 JAL13515.1 GDIQ01164171 GDIQ01063693 JAN31044.1 GECZ01024262 JAS45507.1 GDIQ01002825 JAN91912.1 GDIP01210377 JAJ13025.1 GDIP01012474 JAM91241.1 GDIQ01147423 JAL04303.1 GDIQ01147422 JAL04304.1 GAKP01006304 JAC52648.1 JXJN01031202 AXCM01003036 CAEY01000644 GDIP01233321 JAI90080.1 CVRI01000035 CRK92665.1 JRES01001347 KNC23509.1 GBXI01017248 JAC97043.1 GDIP01212380 JAJ11022.1 AJVK01017655 AJVK01017656 AJVK01017657 AJVK01017658 AJVK01017659 NCKU01000934 RWS13620.1 GEDC01012383 JAS24915.1 CCAG010015251 CCAG010015252 CCAG010015253 CCAG010015254 ATLV01018612 ATLV01018613 KE525239 KFB43260.1 OUUW01000002 SPP77499.1 APCN01000156 APCN01000157 CH379070 EAL30207.3 CH479244 EDW37741.1 GDIP01227769 JAI95632.1 AAAB01008807 EAU77664.2 ABJB010764675 DS735578 EEC07207.1 LT716783 SIW59160.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000000673 UP000069272 UP000092461 UP000053825 UP000007819 UP000095300 UP000092443 UP000092460 UP000075883 UP000092445 UP000091820 UP000015104 UP000075920 UP000005203 UP000079169 UP000183832 UP000037069 UP000075884 UP000092462 UP000285301 UP000092444 UP000075901 UP000192223 UP000030765 UP000268350 UP000075903 UP000075840 UP000001819 UP000008744 UP000007062 UP000265020 UP000001555 UP000261540
UP000000673 UP000069272 UP000092461 UP000053825 UP000007819 UP000095300 UP000092443 UP000092460 UP000075883 UP000092445 UP000091820 UP000015104 UP000075920 UP000005203 UP000079169 UP000183832 UP000037069 UP000075884 UP000092462 UP000285301 UP000092444 UP000075901 UP000192223 UP000030765 UP000268350 UP000075903 UP000075840 UP000001819 UP000008744 UP000007062 UP000265020 UP000001555 UP000261540
PRIDE
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
H9JT80
A0A3S2NE06
A0A2W1BX89
A0A2A4JNB5
A0A2H1VE17
A0A212F2X8
+ More
A0A194RG63 A0A194QDW1 I4DS55 A0A146LTY7 W5JG68 A0A0A9Z4I1 A0A182FGX4 A0A1B0EW48 A0A1L8E3H7 A0A2S2QVR8 A0A2H8TJJ8 A0A0L7RIJ2 J9L3Y9 A0A2S2P1B5 A0A2H8TZW1 A0A147BAL4 A0A1I8NWL6 A0A0P5KJM5 A0A0N8CGS5 A0A0N8DG88 A0A2H8TCQ5 A0A0P5TCN8 A0A1B6LM22 A0A0P5J843 A0A1Z5KY15 A0A0P5C6R8 A0A0N8BJI2 A0A0P6GEZ0 A0A0P5JDV0 A0A0P5JFU2 A0A0P5D4V7 A0A0P6FL89 A0A0P5L055 A0A0P5K3C2 A0A0P5WTG3 A0A0P5WYN8 A0A0P5NZA8 A0A0P4YQE0 A0A0N8BSL2 A0A1B6IHR8 A0A0P5Q670 A0A0P5CJD5 A0A0P5RIX1 A0A0P5JM69 A0A0P5IZU0 A0A0N8BWF6 A0A0P6F9D2 A0A1B6F5Q1 A0A0P6INH2 A0A0N7ZUU5 A0A0P6BNH8 A0A0P5N3U6 A0A0P5NM38 A0A034WFV6 A0A1A9XR29 A0A1B0ALJ4 A0A182MSU3 A0A1A9Z950 A0A1A9W4W3 T1KWH2 A0A182WLH1 A0A0N7ZLM7 A0A088AHR5 A0A1S3D240 A0A1J1I2R7 A0A0L0BWA0 A0A182NK73 A0A0A1WDH0 A0A0P4ZSF2 A0A1B0DNQ4 A0A443RED3 A0A1B6DH23 A0A1B0FM61 A0A182SRU4 A0A1W4WPE5 A0A1W4WPF3 A0A084VZ66 A0A3B0JUW6 A0A1W4X074 A0A2C9H5T1 A0A2C9GPT8 A0A1W4X0F6 Q29E55 B4HAV9 A0A0N7ZNM1 A0A1S4GUY5 A0NBJ8 A0A1W4WZ61 A0A3Q2CZ96 B7PKT5 A0A3B3RBE4 A0A330IZ60
A0A194RG63 A0A194QDW1 I4DS55 A0A146LTY7 W5JG68 A0A0A9Z4I1 A0A182FGX4 A0A1B0EW48 A0A1L8E3H7 A0A2S2QVR8 A0A2H8TJJ8 A0A0L7RIJ2 J9L3Y9 A0A2S2P1B5 A0A2H8TZW1 A0A147BAL4 A0A1I8NWL6 A0A0P5KJM5 A0A0N8CGS5 A0A0N8DG88 A0A2H8TCQ5 A0A0P5TCN8 A0A1B6LM22 A0A0P5J843 A0A1Z5KY15 A0A0P5C6R8 A0A0N8BJI2 A0A0P6GEZ0 A0A0P5JDV0 A0A0P5JFU2 A0A0P5D4V7 A0A0P6FL89 A0A0P5L055 A0A0P5K3C2 A0A0P5WTG3 A0A0P5WYN8 A0A0P5NZA8 A0A0P4YQE0 A0A0N8BSL2 A0A1B6IHR8 A0A0P5Q670 A0A0P5CJD5 A0A0P5RIX1 A0A0P5JM69 A0A0P5IZU0 A0A0N8BWF6 A0A0P6F9D2 A0A1B6F5Q1 A0A0P6INH2 A0A0N7ZUU5 A0A0P6BNH8 A0A0P5N3U6 A0A0P5NM38 A0A034WFV6 A0A1A9XR29 A0A1B0ALJ4 A0A182MSU3 A0A1A9Z950 A0A1A9W4W3 T1KWH2 A0A182WLH1 A0A0N7ZLM7 A0A088AHR5 A0A1S3D240 A0A1J1I2R7 A0A0L0BWA0 A0A182NK73 A0A0A1WDH0 A0A0P4ZSF2 A0A1B0DNQ4 A0A443RED3 A0A1B6DH23 A0A1B0FM61 A0A182SRU4 A0A1W4WPE5 A0A1W4WPF3 A0A084VZ66 A0A3B0JUW6 A0A1W4X074 A0A2C9H5T1 A0A2C9GPT8 A0A1W4X0F6 Q29E55 B4HAV9 A0A0N7ZNM1 A0A1S4GUY5 A0NBJ8 A0A1W4WZ61 A0A3Q2CZ96 B7PKT5 A0A3B3RBE4 A0A330IZ60
PDB
2KV4
E-value=3.1074e-06,
Score=116
Ontologies
PATHWAY
GO
GO:0007399
GO:0005102
GO:0016021
GO:0008355
GO:0007479
GO:0005154
GO:0005615
GO:0048018
GO:0007494
GO:0008284
GO:1903688
GO:0007420
GO:0007173
GO:0035310
GO:2000736
GO:0070374
GO:0030031
GO:0007422
GO:0007482
GO:0008586
GO:0007477
GO:0008201
GO:0035556
GO:0031430
GO:0030017
GO:0031674
GO:0051015
GO:0005515
GO:0046872
GO:0020037
GO:0003676
GO:0015074
GO:0030429
GO:0004003
GO:0005634
GO:0006289
GO:0016286
PANTHER
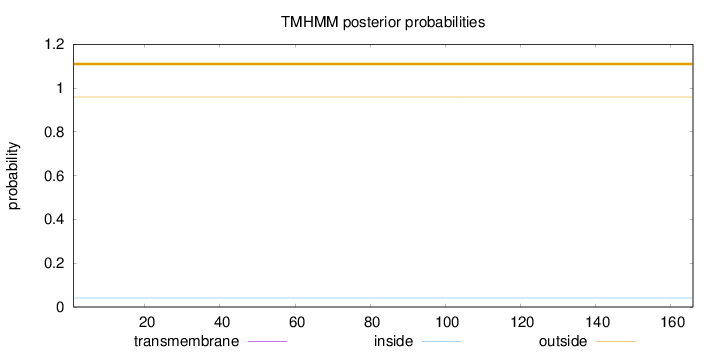
Topology
Length:
166
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0013
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.04099
outside
1 - 166
Population Genetic Test Statistics
Pi
189.736697
Theta
196.111484
Tajima's D
-0.538081
CLR
0.89853
CSRT
0.238038098095095
Interpretation
Uncertain
