Gene
KWMTBOMO12814
Pre Gene Modal
BGIBMGA012741
Annotation
PREDICTED:_uncharacterized_protein_LOC101744504_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.601
Sequence
CDS
ATGACAATGGACGTTCAAAATAATTATCAAAACTATAAAGAATCTTCGCCCGATCCAAAAGAGTCGAATTTGAATATCGAAAGTGCCAGAACGTCTCAGGCCGGGAGAGCTGAGATGTCGACCATCGTAAATCAAGGAAAGAGTTCGAAACGATCATTCGACGTCGCCTTCCTGATGATGCCGGATGATAAATTTCGACAGAAGCAGCCAGAGAAGCAACTGCGAGTGTCCAAGCAAATCTTCCACGGTGAAGAGTGGGAACAATTTCCGAAAAGAATCCCAGAGGCTTATAAACGCGATGAATACTCCAGCGATAACTCCAACACACTAAAAGATCAGGAAGAGGACAGGATTCGAGCGAACAATAACATATCGCCAACTTTTAGTTCCGATATTAACATTGATGTCGGAACGGACGAATACCCCAGTAAGTCGATGTATTGTAGTGATTCAGAAAATTCGGACAAGTCTCGAAGTCGACCTAACTCAAACGAAAGCGCAGCGCGACAGCCTAAATTCCTTCCGGAATACAAAAACAAAATATTTGATGATCCAACACTAATCACGATGCAATCCCGCGCAAGATACGACCGAGCTGAAGAGTCACCTAAAAGTGCTTTTACGAAAGTGTCTCATTATTCTATGAGGTCTCCGAATCCTTCCGTGAGCCCTGATAACCTGCTTTATCAGAATTCCGTCAGCCCTCCGCTGTCCGCGGCCAGTCCACCTGTAAATTTCACCAAGACGCCGCACTTCAACAACCTGCTGAATCCAGCTCATCTGATGAACGGGAGCGGCTTTTTCAAAGGTCAGAACACGCCCTCGTCGTCTCCAAGCTACCCGGATTCAGCATTGCAACAACGTCCAGCCACATTTAGTAAAGCTATTGAGTTTCACGAAAAATCTTCGGATCATAAAGCGTCACCAAATAAGATGAATTTTCTACCGTTCAGACCCGAAATACCGTATCTTCAAGCCGGAGGGTCCTATCAATTTCAGAACTTCCAAACGGCAAATTCGGATTTCATGAAATTTCCTGTGCCTCCTCGAGAGCCGGTCAGTCCGCTGATAGCAAACCCAGCAGCAGCCATCCTGAGTACTTTACTGCCGCCTACTTTAGCTGCTTTTTCGTTGCCAGCACAGAATGTTTGCGCTAAGTGCAGCATCAGTTTCAGGATGACTTCTGACTTAGTTTACCACATGCGAACGCATCACAAGAGCGAGAACGTAGTCGATCCCAATAGGAGAAAAAGAGAGGAGAAACTGAAGTGTCCGGTTTGCAATGAGAGCTTCAGAGAGAGGCATCATTTAACAAGACACATGACGGCCCACCAGGACAAAGAAGGCGACATGGACGACGTGTCCCCTTCGCAAAATTACAAGAAATCCAAACAATTTTATTCCCACAATAGTGGATCGATAATTCACAAGTAA
Protein
MTMDVQNNYQNYKESSPDPKESNLNIESARTSQAGRAEMSTIVNQGKSSKRSFDVAFLMMPDDKFRQKQPEKQLRVSKQIFHGEEWEQFPKRIPEAYKRDEYSSDNSNTLKDQEEDRIRANNNISPTFSSDINIDVGTDEYPSKSMYCSDSENSDKSRSRPNSNESAARQPKFLPEYKNKIFDDPTLITMQSRARYDRAEESPKSAFTKVSHYSMRSPNPSVSPDNLLYQNSVSPPLSAASPPVNFTKTPHFNNLLNPAHLMNGSGFFKGQNTPSSSPSYPDSALQQRPATFSKAIEFHEKSSDHKASPNKMNFLPFRPEIPYLQAGGSYQFQNFQTANSDFMKFPVPPREPVSPLIANPAAAILSTLLPPTLAAFSLPAQNVCAKCSISFRMTSDLVYHMRTHHKSENVVDPNRRKREEKLKCPVCNESFRERHHLTRHMTAHQDKEGDMDDVSPSQNYKKSKQFYSHNSGSIIHK
Summary
Uniprot
Pubmed
EMBL
BABH01034766
KZ149907
PZC78262.1
NWSH01001320
PCG71709.1
ODYU01001331
+ More
SOQ37369.1 RSAL01000176 RVE45042.1 AGBW02012596 OWR44737.1 KQ460416 KPJ14925.1 KQ459324 KPJ01649.1 JTDY01006512 KOB65950.1 KQ971343 EFA03154.2 GL768061 EFZ12666.1 KQ981387 KYN41998.1 KQ980204 KYN17273.1 LBMM01000599 KMQ97985.1 KQ977971 KYM98405.1 ADTU01015404 GL888673 EGI58569.1 KQ976574 KYM80124.1 KQ982294 KYQ58316.1 KQ435727 KOX77872.1
SOQ37369.1 RSAL01000176 RVE45042.1 AGBW02012596 OWR44737.1 KQ460416 KPJ14925.1 KQ459324 KPJ01649.1 JTDY01006512 KOB65950.1 KQ971343 EFA03154.2 GL768061 EFZ12666.1 KQ981387 KYN41998.1 KQ980204 KYN17273.1 LBMM01000599 KMQ97985.1 KQ977971 KYM98405.1 ADTU01015404 GL888673 EGI58569.1 KQ976574 KYM80124.1 KQ982294 KYQ58316.1 KQ435727 KOX77872.1
Proteomes
PRIDE
Pfam
PF00096 zf-C2H2
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
PDB
1MEY
E-value=0.0123283,
Score=92
Ontologies
GO
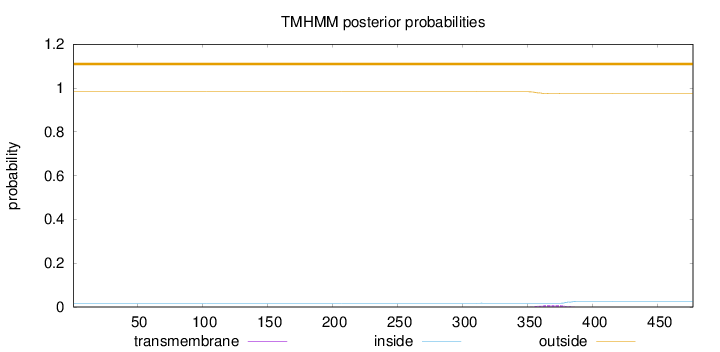
Topology
Length:
477
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19077
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01747
outside
1 - 477
Population Genetic Test Statistics
Pi
133.562186
Theta
169.013299
Tajima's D
-0.657462
CLR
0.011079
CSRT
0.19954002299885
Interpretation
Uncertain
