Gene
KWMTBOMO12813
Pre Gene Modal
BGIBMGA012735
Annotation
PREDICTED:_netrin_receptor_UNC5C-like_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.802
Sequence
CDS
ATGATTACAATATTTATTGGCGAAGTAAAAAACGGAGATCCAGAACCAACGACAGCAAAAGAACTAAGACCAGAGTACATACATCCAGATGGCCTACTAACCTTCGACCCTTACCTTTCTGACATAAAACCTCAAGAGGAAGAAGAAGAAGACGATTATATACCTTTACCAGAAAACGATGAAACCATTGATGGTCTTCCAATATTCTTATTAGAACCTAAAAACTCGTATGTCATTAGGAATAAACCAGCCACGATTCTCTGTAGGGCTGCCAATGCTCTTCAAGTGTTCTTCAAATGCAATGACGTGAGAACTGATAAGACAGTACAATACGAACACGTGGATCCTCAGAATGGAGTGAGGGTGGTCGAAGCAGAGCTAAATGTGACCAGAAATGAGTTAGATGAATATTTCGGTGGAAAATACGGGTGTGAGTGCTACGCTTGGAATAGTAAAGGGCGGATTCGCAGTCAGTCCATTTTCATCGAATACGCTTTCATCAAGAAGCATTTCGTGCAGCAACCGCAGTCAGCGACAGTCGAATCAGGCAAGAGCGTCACATTCCGATGTATTCCGCCACAAGCCACACCAGCTCCAAGTCTCAAATGGACAAGAAATGGAGTGCCTATTGAGGCAAAGGACGAAAATTTAGTGCTTCCTAATGTTGGGTTGCAGGACATGACAAACTACACCTGTATAGCTGAGAACGTCGCCGGTCGGCGCGAGTCGAACGTGGCCGTGCTCTCTGTATACGTAAATGGAGGCTGGTCAGCCTGGTCTCCGTGGATGCCGTGTCGTTGCGGCTCCAGAATTAGCGGAAGACGTCGCACGAGAACTTGCACAGAGCCTCCGCCGGCCAACGGAGGAGCGCCCTGCAGAGGCCAGGCCTCGCAGAGCGACGAGGAATGCTTACGATGTCAGACCGAAGGTTCATGGTCTCCGTGGAGTGCCTGGTCCACTTGTAGTGGAGACTGCGTCAGGGTCCGTCGGAGGGAGTGCACCGGAGAGTGTACCGGGTCTACCTTGCAACATGCCACTTGCGTCGATGGCATGTGTCCCGCTGGCCTGAGGCTGTATAACACTAACGACGTCACTATGTACGTGGGAGTTGGCATCACGGTGGCGATGCTGGTGGCGGGAGCTACCATCCTCTACGTCTGGTGCAGATTTAGGAGCAGGCCCGGCTACACCGCCGCCAGGGCAGGTACTCCAAAACACATCGGTCGAGATAAAGGTAACGCAGGACCGGACTTGACGAGGGCCTGCAACGAGTATTGGATGCAGGGGCCCGACAATCATTACGACATGCCTCACGTGAGAGACAGTTATTCTACCCCGTTTGGAAACAGAAGTAGACATCAGTCGTCCATGGGAAGTAATCACTACGAGATGAAACCGTACAGCCCTTCGGGCGAATCCGCCTCTAGCTGCTACACAAATTCCCGAACAGTGACCTCGAGGTCCAGTGAATGTTCCTCATCCCAATTAGCAGATTTGGTGACCGGAACCCCCACCCCGGTGTCCAAGGTCGACGTGGCGGTCACCATGCCGACCAGCCACACAGACAACAGCTACAGAGACAAAATATGCTTGACCATAGTTAAATAG
Protein
MITIFIGEVKNGDPEPTTAKELRPEYIHPDGLLTFDPYLSDIKPQEEEEEDDYIPLPENDETIDGLPIFLLEPKNSYVIRNKPATILCRAANALQVFFKCNDVRTDKTVQYEHVDPQNGVRVVEAELNVTRNELDEYFGGKYGCECYAWNSKGRIRSQSIFIEYAFIKKHFVQQPQSATVESGKSVTFRCIPPQATPAPSLKWTRNGVPIEAKDENLVLPNVGLQDMTNYTCIAENVAGRRESNVAVLSVYVNGGWSAWSPWMPCRCGSRISGRRRTRTCTEPPPANGGAPCRGQASQSDEECLRCQTEGSWSPWSAWSTCSGDCVRVRRRECTGECTGSTLQHATCVDGMCPAGLRLYNTNDVTMYVGVGITVAMLVAGATILYVWCRFRSRPGYTAARAGTPKHIGRDKGNAGPDLTRACNEYWMQGPDNHYDMPHVRDSYSTPFGNRSRHQSSMGSNHYEMKPYSPSGESASSCYTNSRTVTSRSSECSSSQLADLVTGTPTPVSKVDVAVTMPTSHTDNSYRDKICLTIVK
Summary
Similarity
Belongs to the serpin family.
Uniprot
A0A2W1BV79
A0A194RBR7
H9JT73
A0A2A4JJ15
A0A2H1V962
A0A3S2TN21
+ More
A0A194Q7Z8 A0A0L7KYD2 A0A212EVY5 A0A437BJV8 A0A194Q8C5 A0A2A4JM37 A0A2A4JLB3 A0A2W1BXA1 A0A154PEH5 A0A0K8TDJ2 A0A1Y1KGW7 A0A146LCK0 A0A0A9VZT2 A0A0N1I685 A0A195CJV7 A0A2A4IZN1 A0A194PXF9 A0A2W1BMT9 A0A067RP65 A0A1B6BY64 A0A1B6KVK9 A0A1B6LH58 A0A0L7RHT7 W8B2P2 W8AYM5 A0A0A1X6V4 A0A088A971 A0A034W4Y3 A0A158NTE6 A0A2S2PSX4 A0A0K8UWV4 A0A3B3YP05 A0A3B5RDJ2 A0A3B3XAI2 A0A3B3VM99 A0A151X1U2 A0A3P9Q5U0 A0A3P9Q5P4 F4W406 A0A146ZII3 A0A2H8TRB2 A0A2I4D0E9 A0A437D086 A0A3Q3BCL4 A0A026VUN0 A0A2I4D0E5 H2LLZ0 H2LLY9 A0A195B423 A0A026VX76 A0A0M9A636 A0A1A7ZTP1 A0A1A8Q8H3 A0A1A8C1H1 A0A1A8V5D7 A0A3Q0R8V6 A0A3Q3JBA1 A0A3B3XAX3 A0A3Q1IIL7 A0A087YKP9 J9KU90 M4ARZ4 A0A3B3YP00 A0A3P9Q675 A0A3B4YRF8 G3NYB5 A0A3Q1IQU9 A0A3Q2WVM9 A0A3B4FZ93 A0A3P8QJH3 A0A3B4VIA3 A0A3Q4H2S8 E2APQ4 A0A3Q3MXJ1 A0A3P8YXM7 A0A2S2PVW1 A0A146UWS2 Q4W5H4 U5EYZ9 A0A3B4XEP7 A0A3Q3BCI7 A0A195DVW8 A0A3B3H6L5
A0A194Q7Z8 A0A0L7KYD2 A0A212EVY5 A0A437BJV8 A0A194Q8C5 A0A2A4JM37 A0A2A4JLB3 A0A2W1BXA1 A0A154PEH5 A0A0K8TDJ2 A0A1Y1KGW7 A0A146LCK0 A0A0A9VZT2 A0A0N1I685 A0A195CJV7 A0A2A4IZN1 A0A194PXF9 A0A2W1BMT9 A0A067RP65 A0A1B6BY64 A0A1B6KVK9 A0A1B6LH58 A0A0L7RHT7 W8B2P2 W8AYM5 A0A0A1X6V4 A0A088A971 A0A034W4Y3 A0A158NTE6 A0A2S2PSX4 A0A0K8UWV4 A0A3B3YP05 A0A3B5RDJ2 A0A3B3XAI2 A0A3B3VM99 A0A151X1U2 A0A3P9Q5U0 A0A3P9Q5P4 F4W406 A0A146ZII3 A0A2H8TRB2 A0A2I4D0E9 A0A437D086 A0A3Q3BCL4 A0A026VUN0 A0A2I4D0E5 H2LLZ0 H2LLY9 A0A195B423 A0A026VX76 A0A0M9A636 A0A1A7ZTP1 A0A1A8Q8H3 A0A1A8C1H1 A0A1A8V5D7 A0A3Q0R8V6 A0A3Q3JBA1 A0A3B3XAX3 A0A3Q1IIL7 A0A087YKP9 J9KU90 M4ARZ4 A0A3B3YP00 A0A3P9Q675 A0A3B4YRF8 G3NYB5 A0A3Q1IQU9 A0A3Q2WVM9 A0A3B4FZ93 A0A3P8QJH3 A0A3B4VIA3 A0A3Q4H2S8 E2APQ4 A0A3Q3MXJ1 A0A3P8YXM7 A0A2S2PVW1 A0A146UWS2 Q4W5H4 U5EYZ9 A0A3B4XEP7 A0A3Q3BCI7 A0A195DVW8 A0A3B3H6L5
Pubmed
EMBL
KZ149907
PZC78261.1
KQ460416
KPJ14924.1
BABH01034762
NWSH01001320
+ More
PCG71708.1 ODYU01001331 SOQ37368.1 RSAL01000046 RVE50542.1 KQ459324 KPJ01648.1 JTDY01004517 KOB68059.1 AGBW02012107 OWR45646.1 RVE50538.1 KPJ01644.1 NWSH01001062 PCG72846.1 PCG72847.1 PZC78254.1 KQ434870 KZC09610.1 GBRD01002257 JAG63564.1 GEZM01086422 JAV59460.1 GDHC01018809 GDHC01013807 JAP99819.1 JAQ04822.1 GBHO01043896 JAF99707.1 KQ460874 KPJ11666.1 KQ977642 KYN01011.1 NWSH01004342 PCG65211.1 KQ459586 KPI98031.1 KZ150134 PZC73003.1 KK852508 KDR22415.1 GEDC01031076 JAS06222.1 GEBQ01024488 JAT15489.1 GEBQ01016935 JAT23042.1 KQ414585 KOC70552.1 GAMC01015247 JAB91308.1 GAMC01015248 JAB91307.1 GBXI01007470 JAD06822.1 GAKP01009178 GAKP01009177 GAKP01009176 JAC49774.1 ADTU01025884 ADTU01025885 GGMR01019327 MBY31946.1 GDHF01025673 GDHF01021499 JAI26641.1 JAI30815.1 KQ982585 KYQ54236.1 GL887491 EGI71002.1 GCES01020152 JAR66171.1 GFXV01004337 MBW16142.1 CM012445 RVE68149.1 KK107847 EZA47457.1 KQ976625 KYM78959.1 EZA47459.1 KQ435727 KOX77995.1 HADY01007659 SBP46144.1 HAEH01003003 HAEI01004676 SBR89826.1 HADZ01008765 SBP72706.1 HAEJ01014574 SBS55031.1 AYCK01002916 AYCK01002917 AYCK01002918 AYCK01002919 ABLF02041018 GL441572 EFN64605.1 GGMS01000443 MBY69646.1 GCES01076885 JAR09438.1 AC098584 AAY41022.1 GANO01001690 JAB58181.1 KQ980295 KYN16887.1
PCG71708.1 ODYU01001331 SOQ37368.1 RSAL01000046 RVE50542.1 KQ459324 KPJ01648.1 JTDY01004517 KOB68059.1 AGBW02012107 OWR45646.1 RVE50538.1 KPJ01644.1 NWSH01001062 PCG72846.1 PCG72847.1 PZC78254.1 KQ434870 KZC09610.1 GBRD01002257 JAG63564.1 GEZM01086422 JAV59460.1 GDHC01018809 GDHC01013807 JAP99819.1 JAQ04822.1 GBHO01043896 JAF99707.1 KQ460874 KPJ11666.1 KQ977642 KYN01011.1 NWSH01004342 PCG65211.1 KQ459586 KPI98031.1 KZ150134 PZC73003.1 KK852508 KDR22415.1 GEDC01031076 JAS06222.1 GEBQ01024488 JAT15489.1 GEBQ01016935 JAT23042.1 KQ414585 KOC70552.1 GAMC01015247 JAB91308.1 GAMC01015248 JAB91307.1 GBXI01007470 JAD06822.1 GAKP01009178 GAKP01009177 GAKP01009176 JAC49774.1 ADTU01025884 ADTU01025885 GGMR01019327 MBY31946.1 GDHF01025673 GDHF01021499 JAI26641.1 JAI30815.1 KQ982585 KYQ54236.1 GL887491 EGI71002.1 GCES01020152 JAR66171.1 GFXV01004337 MBW16142.1 CM012445 RVE68149.1 KK107847 EZA47457.1 KQ976625 KYM78959.1 EZA47459.1 KQ435727 KOX77995.1 HADY01007659 SBP46144.1 HAEH01003003 HAEI01004676 SBR89826.1 HADZ01008765 SBP72706.1 HAEJ01014574 SBS55031.1 AYCK01002916 AYCK01002917 AYCK01002918 AYCK01002919 ABLF02041018 GL441572 EFN64605.1 GGMS01000443 MBY69646.1 GCES01076885 JAR09438.1 AC098584 AAY41022.1 GANO01001690 JAB58181.1 KQ980295 KYN16887.1
Proteomes
UP000053240
UP000005204
UP000218220
UP000283053
UP000053268
UP000037510
+ More
UP000007151 UP000076502 UP000078542 UP000027135 UP000053825 UP000005203 UP000005205 UP000261480 UP000002852 UP000261500 UP000075809 UP000242638 UP000007755 UP000192220 UP000264800 UP000053097 UP000001038 UP000078540 UP000053105 UP000261340 UP000261600 UP000265040 UP000028760 UP000007819 UP000261360 UP000007635 UP000264840 UP000261460 UP000265100 UP000261420 UP000261580 UP000000311 UP000261640 UP000265140 UP000078492
UP000007151 UP000076502 UP000078542 UP000027135 UP000053825 UP000005203 UP000005205 UP000261480 UP000002852 UP000261500 UP000075809 UP000242638 UP000007755 UP000192220 UP000264800 UP000053097 UP000001038 UP000078540 UP000053105 UP000261340 UP000261600 UP000265040 UP000028760 UP000007819 UP000261360 UP000007635 UP000264840 UP000261460 UP000265100 UP000261420 UP000261580 UP000000311 UP000261640 UP000265140 UP000078492
Interpro
IPR013783
Ig-like_fold
+ More
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR036383 TSP1_rpt_sf
IPR037936 UNC5
IPR000884 TSP1_rpt
IPR011029 DEATH-like_dom_sf
IPR000906 ZU5_dom
IPR000488 Death_domain
IPR033772 UPA
IPR013098 Ig_I-set
IPR042185 Serpin_sf_2
IPR042178 Serpin_sf_1
IPR023796 Serpin_dom
IPR036186 Serpin_sf
IPR042154 Death_UNC5C
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR036383 TSP1_rpt_sf
IPR037936 UNC5
IPR000884 TSP1_rpt
IPR011029 DEATH-like_dom_sf
IPR000906 ZU5_dom
IPR000488 Death_domain
IPR033772 UPA
IPR013098 Ig_I-set
IPR042185 Serpin_sf_2
IPR042178 Serpin_sf_1
IPR023796 Serpin_dom
IPR036186 Serpin_sf
IPR042154 Death_UNC5C
Gene 3D
ProteinModelPortal
A0A2W1BV79
A0A194RBR7
H9JT73
A0A2A4JJ15
A0A2H1V962
A0A3S2TN21
+ More
A0A194Q7Z8 A0A0L7KYD2 A0A212EVY5 A0A437BJV8 A0A194Q8C5 A0A2A4JM37 A0A2A4JLB3 A0A2W1BXA1 A0A154PEH5 A0A0K8TDJ2 A0A1Y1KGW7 A0A146LCK0 A0A0A9VZT2 A0A0N1I685 A0A195CJV7 A0A2A4IZN1 A0A194PXF9 A0A2W1BMT9 A0A067RP65 A0A1B6BY64 A0A1B6KVK9 A0A1B6LH58 A0A0L7RHT7 W8B2P2 W8AYM5 A0A0A1X6V4 A0A088A971 A0A034W4Y3 A0A158NTE6 A0A2S2PSX4 A0A0K8UWV4 A0A3B3YP05 A0A3B5RDJ2 A0A3B3XAI2 A0A3B3VM99 A0A151X1U2 A0A3P9Q5U0 A0A3P9Q5P4 F4W406 A0A146ZII3 A0A2H8TRB2 A0A2I4D0E9 A0A437D086 A0A3Q3BCL4 A0A026VUN0 A0A2I4D0E5 H2LLZ0 H2LLY9 A0A195B423 A0A026VX76 A0A0M9A636 A0A1A7ZTP1 A0A1A8Q8H3 A0A1A8C1H1 A0A1A8V5D7 A0A3Q0R8V6 A0A3Q3JBA1 A0A3B3XAX3 A0A3Q1IIL7 A0A087YKP9 J9KU90 M4ARZ4 A0A3B3YP00 A0A3P9Q675 A0A3B4YRF8 G3NYB5 A0A3Q1IQU9 A0A3Q2WVM9 A0A3B4FZ93 A0A3P8QJH3 A0A3B4VIA3 A0A3Q4H2S8 E2APQ4 A0A3Q3MXJ1 A0A3P8YXM7 A0A2S2PVW1 A0A146UWS2 Q4W5H4 U5EYZ9 A0A3B4XEP7 A0A3Q3BCI7 A0A195DVW8 A0A3B3H6L5
A0A194Q7Z8 A0A0L7KYD2 A0A212EVY5 A0A437BJV8 A0A194Q8C5 A0A2A4JM37 A0A2A4JLB3 A0A2W1BXA1 A0A154PEH5 A0A0K8TDJ2 A0A1Y1KGW7 A0A146LCK0 A0A0A9VZT2 A0A0N1I685 A0A195CJV7 A0A2A4IZN1 A0A194PXF9 A0A2W1BMT9 A0A067RP65 A0A1B6BY64 A0A1B6KVK9 A0A1B6LH58 A0A0L7RHT7 W8B2P2 W8AYM5 A0A0A1X6V4 A0A088A971 A0A034W4Y3 A0A158NTE6 A0A2S2PSX4 A0A0K8UWV4 A0A3B3YP05 A0A3B5RDJ2 A0A3B3XAI2 A0A3B3VM99 A0A151X1U2 A0A3P9Q5U0 A0A3P9Q5P4 F4W406 A0A146ZII3 A0A2H8TRB2 A0A2I4D0E9 A0A437D086 A0A3Q3BCL4 A0A026VUN0 A0A2I4D0E5 H2LLZ0 H2LLY9 A0A195B423 A0A026VX76 A0A0M9A636 A0A1A7ZTP1 A0A1A8Q8H3 A0A1A8C1H1 A0A1A8V5D7 A0A3Q0R8V6 A0A3Q3JBA1 A0A3B3XAX3 A0A3Q1IIL7 A0A087YKP9 J9KU90 M4ARZ4 A0A3B3YP00 A0A3P9Q675 A0A3B4YRF8 G3NYB5 A0A3Q1IQU9 A0A3Q2WVM9 A0A3B4FZ93 A0A3P8QJH3 A0A3B4VIA3 A0A3Q4H2S8 E2APQ4 A0A3Q3MXJ1 A0A3P8YXM7 A0A2S2PVW1 A0A146UWS2 Q4W5H4 U5EYZ9 A0A3B4XEP7 A0A3Q3BCI7 A0A195DVW8 A0A3B3H6L5
PDB
5FTT
E-value=2.96662e-39,
Score=408
Ontologies
GO
PANTHER
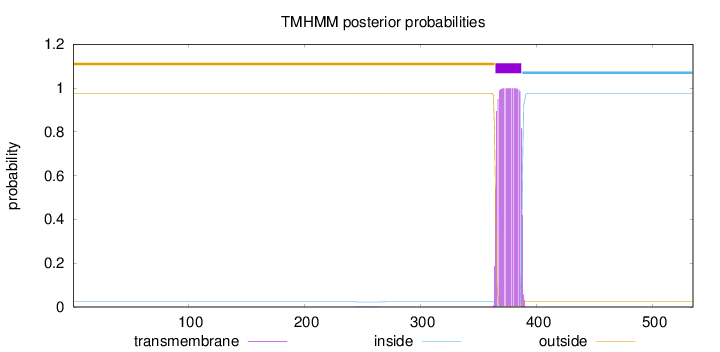
Topology
Length:
535
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.89196
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02440
outside
1 - 364
TMhelix
365 - 387
inside
388 - 535
Population Genetic Test Statistics
Pi
300.959487
Theta
189.827332
Tajima's D
2.862379
CLR
10.402839
CSRT
0.972751362431878
Interpretation
Uncertain
