Gene
KWMTBOMO12811
Pre Gene Modal
BGIBMGA012737
Annotation
PREDICTED:_uncharacterized_protein_LOC105841592_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.208
Sequence
CDS
ATGACGAACGATCTAGCGAAGAATCGACATAGCGGCTTCCAATCCTATATCAAATACAGAGCCTATTGTTTTGGAGACAATATCGCTTCCTTTGATGATCTAAAGGGTATTATTGAAAAAGAGAGAATCGAGGCGTTAAAAGATGTTTATGCAAACGTTGCCGACATAGATTTGTTGGCTGGAGTTTGGGTGGAGATCTTCAAAGAAGGATCTCATGTTCCGCCTACATTCTACTGTATAGTCAAAGATCAGCTGCTTCATAATATAAAGTCCGATAGACATTGGTATGAGCGAGAAAAAAGACCTAATGCTTTTACTGAGGCACAACTAGCTGAAATACGGAAGGTAACTGGGGCTCGGATACTATGTGATATTGGTGATTCGGTTACAGCAATACAGCCTTCAGTCTTCAAGCTGCCAGGAGAAGGAAATAAAATTGTGGACTGCTCCGAAATACCGGATATTGACTACACAGCCTGGTCAGATAATTAG
Protein
MTNDLAKNRHSGFQSYIKYRAYCFGDNIASFDDLKGIIEKERIEALKDVYANVADIDLLAGVWVEIFKEGSHVPPTFYCIVKDQLLHNIKSDRHWYEREKRPNAFTEAQLAEIRKVTGARILCDIGDSVTAIQPSVFKLPGEGNKIVDCSEIPDIDYTAWSDN
Summary
Uniprot
H9JT75
A0A2W1BT77
A0A088MGW5
A0A2A4JI21
H9JT76
A0A2H1WGR9
+ More
A0A2H1W8U7 I4DS54 I4DN32 A0A194Q7Y0 I4DJ10 A0A194RBC8 A0A194Q9H2 A0A194RAP6 A0A194QDV6 A0A212ETD1 A0A1E1WUQ6 A0A2H1W9U4 A0A2W1BYE5 A0A2A4IUC0 H9ISL2 A0A2W1BT67 A0A3S2NLC3 A0A2A4J7D3 A0A2H1W4U4 A0A3S2LP82 A0A2H1WBG2 A0A2H1VUX0 A0A437AS20 A0A437ASV3 A0A2H1V5L5 O96645 Q177U0 Q16E76 A0A437ARC6 A0A1W4XPE3 W8C2M9 H9JQT0 A0A0P6H2D0 A0A0M9AC65 A0A182G7N2 A0A1B6I2F8 A0A1W6EVW7 E2BYM5 A0A1B6KS40 A0A0P6FAZ7 A0A1B6CGS3 A0A1B6FKB9 A0A0N8E1B0 A0A437AYB0 A0A1B6ME87 A0A1B6D0W8 A0A0P5RF89 A0A182T872 A0A182KBY5 A0A0N8EAV2 A0A0P6DND4 A0A0P6DE26 A0A0P6DDQ5 A0A0P6DGA0 A0A0P6GA01 A0A0P5PP69 A0A0P6FNT6 A0A0P5QCR3 A0A0P6E2J6 D6WB23 A0A0P6E456 A0A0P6F2H6 B0XE84 B0WM28 A0A139WNI1 A0A182Q218 A0A182VL61 A0A182KV49 A0A182WXU8 A0A1S4H4U5 A0A182TM70 A0A182I5W9 Q7QDZ3 A0A0K8VYA7 A0A0P4ZAS8 A0A0P6FX89 A0A0P4ZCC9 H9JT78 A0A0P5JNG9 A0A0P5SA74 A0A0P5M0T2 A0A0N8CDW0 A0A2J7R3R2 A0A0N8BFF0 A0A0P5SIR0 A0A0P6GB44 A0A0P6FJQ0 A0A0P6DU56 A0A164VLV1 A0A0P5VUV2 A0A0P5NKS6 A0A0P5WBR2 A0A0P5SKA9
A0A2H1W8U7 I4DS54 I4DN32 A0A194Q7Y0 I4DJ10 A0A194RBC8 A0A194Q9H2 A0A194RAP6 A0A194QDV6 A0A212ETD1 A0A1E1WUQ6 A0A2H1W9U4 A0A2W1BYE5 A0A2A4IUC0 H9ISL2 A0A2W1BT67 A0A3S2NLC3 A0A2A4J7D3 A0A2H1W4U4 A0A3S2LP82 A0A2H1WBG2 A0A2H1VUX0 A0A437AS20 A0A437ASV3 A0A2H1V5L5 O96645 Q177U0 Q16E76 A0A437ARC6 A0A1W4XPE3 W8C2M9 H9JQT0 A0A0P6H2D0 A0A0M9AC65 A0A182G7N2 A0A1B6I2F8 A0A1W6EVW7 E2BYM5 A0A1B6KS40 A0A0P6FAZ7 A0A1B6CGS3 A0A1B6FKB9 A0A0N8E1B0 A0A437AYB0 A0A1B6ME87 A0A1B6D0W8 A0A0P5RF89 A0A182T872 A0A182KBY5 A0A0N8EAV2 A0A0P6DND4 A0A0P6DE26 A0A0P6DDQ5 A0A0P6DGA0 A0A0P6GA01 A0A0P5PP69 A0A0P6FNT6 A0A0P5QCR3 A0A0P6E2J6 D6WB23 A0A0P6E456 A0A0P6F2H6 B0XE84 B0WM28 A0A139WNI1 A0A182Q218 A0A182VL61 A0A182KV49 A0A182WXU8 A0A1S4H4U5 A0A182TM70 A0A182I5W9 Q7QDZ3 A0A0K8VYA7 A0A0P4ZAS8 A0A0P6FX89 A0A0P4ZCC9 H9JT78 A0A0P5JNG9 A0A0P5SA74 A0A0P5M0T2 A0A0N8CDW0 A0A2J7R3R2 A0A0N8BFF0 A0A0P5SIR0 A0A0P6GB44 A0A0P6FJQ0 A0A0P6DU56 A0A164VLV1 A0A0P5VUV2 A0A0P5NKS6 A0A0P5WBR2 A0A0P5SKA9
Pubmed
EMBL
BABH01034751
BABH01034752
BABH01034753
BABH01034754
KZ149907
PZC78259.1
+ More
KJ995811 AIN39494.1 NWSH01001320 PCG71707.1 BABH01034742 ODYU01008556 SOQ52260.1 ODYU01007063 SOQ49501.1 AK405426 BAM20744.1 AK402700 BAM19322.1 KQ459324 KPJ01647.1 AK401278 BAM17900.1 KQ460416 KPJ14922.1 KPJ01645.1 KPJ14923.1 KPJ01646.1 AGBW02012596 OWR44740.1 GDQN01000342 JAT90712.1 ODYU01007226 SOQ49823.1 PZC78257.1 NWSH01006580 PCG63355.1 BABH01013447 BABH01013448 PZC78258.1 RSAL01000046 RVE50539.1 NWSH01002854 PCG67434.1 ODYU01006337 SOQ48101.1 RSAL01001640 RVE40745.1 ODYU01007479 SOQ50286.1 ODYU01004596 SOQ44641.1 RVE40744.1 RSAL01000844 RVE41114.1 ODYU01000771 SOQ36066.1 AF098717 AAC97504.1 CH477371 EAT42428.1 CH478998 EAT32533.1 RSAL01003076 RVE40348.1 GAMC01010556 JAB95999.1 BABH01028746 BABH01028747 BABH01028748 GDIQ01046750 JAN47987.1 KQ435706 KOX80149.1 JXUM01046680 KQ561486 KXJ78429.1 GECU01026590 JAS81116.1 KY563436 ARK19845.1 GL451499 EFN79179.1 GEBQ01025718 JAT14259.1 GDIQ01052795 JAN41942.1 GEDC01024763 JAS12535.1 GECZ01019152 JAS50617.1 GDIQ01071588 JAN23149.1 RSAL01000273 RVE43156.1 GEBQ01005757 JAT34220.1 GEDC01017960 JAS19338.1 GDIQ01101581 JAL50145.1 GDIQ01044852 JAN49885.1 GDIQ01078682 JAN16055.1 GDIQ01078680 JAN16057.1 GDIQ01078681 JAN16056.1 GDIQ01078683 JAN16054.1 GDIQ01046751 JAN47986.1 GDIQ01125477 JAL26249.1 GDIQ01044853 JAN49884.1 GDIQ01125476 JAL26250.1 GDIQ01068751 JAN25986.1 KQ971311 EEZ99334.2 GDIQ01068746 JAN25991.1 GDIQ01068747 JAN25990.1 DS232811 EDS45917.1 DS231994 EDS30856.1 KYB29427.1 AXCN02001762 AAAB01008848 APCN01003535 EAA07042.4 GDHF01011408 GDHF01008719 JAI40906.1 JAI43595.1 GDIP01215167 JAJ08235.1 GDIQ01052794 JAN41943.1 GDIP01215170 JAJ08232.1 BABH01034746 BABH01034747 GDIQ01200602 JAK51123.1 GDIP01142426 JAL61288.1 GDIQ01171340 JAK80385.1 GDIP01141367 JAL62347.1 NEVH01007818 PNF35455.1 GDIQ01183059 JAK68666.1 GDIP01139278 JAL64436.1 GDIQ01035995 JAN58742.1 GDIQ01058737 JAN36000.1 GDIQ01072777 JAN21960.1 LRGB01001361 KZS12450.1 GDIP01109522 JAL94192.1 GDIQ01148117 JAL03609.1 GDIP01089427 JAM14288.1 GDIP01138714 JAL65000.1
KJ995811 AIN39494.1 NWSH01001320 PCG71707.1 BABH01034742 ODYU01008556 SOQ52260.1 ODYU01007063 SOQ49501.1 AK405426 BAM20744.1 AK402700 BAM19322.1 KQ459324 KPJ01647.1 AK401278 BAM17900.1 KQ460416 KPJ14922.1 KPJ01645.1 KPJ14923.1 KPJ01646.1 AGBW02012596 OWR44740.1 GDQN01000342 JAT90712.1 ODYU01007226 SOQ49823.1 PZC78257.1 NWSH01006580 PCG63355.1 BABH01013447 BABH01013448 PZC78258.1 RSAL01000046 RVE50539.1 NWSH01002854 PCG67434.1 ODYU01006337 SOQ48101.1 RSAL01001640 RVE40745.1 ODYU01007479 SOQ50286.1 ODYU01004596 SOQ44641.1 RVE40744.1 RSAL01000844 RVE41114.1 ODYU01000771 SOQ36066.1 AF098717 AAC97504.1 CH477371 EAT42428.1 CH478998 EAT32533.1 RSAL01003076 RVE40348.1 GAMC01010556 JAB95999.1 BABH01028746 BABH01028747 BABH01028748 GDIQ01046750 JAN47987.1 KQ435706 KOX80149.1 JXUM01046680 KQ561486 KXJ78429.1 GECU01026590 JAS81116.1 KY563436 ARK19845.1 GL451499 EFN79179.1 GEBQ01025718 JAT14259.1 GDIQ01052795 JAN41942.1 GEDC01024763 JAS12535.1 GECZ01019152 JAS50617.1 GDIQ01071588 JAN23149.1 RSAL01000273 RVE43156.1 GEBQ01005757 JAT34220.1 GEDC01017960 JAS19338.1 GDIQ01101581 JAL50145.1 GDIQ01044852 JAN49885.1 GDIQ01078682 JAN16055.1 GDIQ01078680 JAN16057.1 GDIQ01078681 JAN16056.1 GDIQ01078683 JAN16054.1 GDIQ01046751 JAN47986.1 GDIQ01125477 JAL26249.1 GDIQ01044853 JAN49884.1 GDIQ01125476 JAL26250.1 GDIQ01068751 JAN25986.1 KQ971311 EEZ99334.2 GDIQ01068746 JAN25991.1 GDIQ01068747 JAN25990.1 DS232811 EDS45917.1 DS231994 EDS30856.1 KYB29427.1 AXCN02001762 AAAB01008848 APCN01003535 EAA07042.4 GDHF01011408 GDHF01008719 JAI40906.1 JAI43595.1 GDIP01215167 JAJ08235.1 GDIQ01052794 JAN41943.1 GDIP01215170 JAJ08232.1 BABH01034746 BABH01034747 GDIQ01200602 JAK51123.1 GDIP01142426 JAL61288.1 GDIQ01171340 JAK80385.1 GDIP01141367 JAL62347.1 NEVH01007818 PNF35455.1 GDIQ01183059 JAK68666.1 GDIP01139278 JAL64436.1 GDIQ01035995 JAN58742.1 GDIQ01058737 JAN36000.1 GDIQ01072777 JAN21960.1 LRGB01001361 KZS12450.1 GDIP01109522 JAL94192.1 GDIQ01148117 JAL03609.1 GDIP01089427 JAM14288.1 GDIP01138714 JAL65000.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000008820 UP000192223 UP000053105 UP000069940 UP000249989 UP000008237 UP000075901 UP000075881 UP000007266 UP000002320 UP000075886 UP000075903 UP000075882 UP000076407 UP000075902 UP000075840 UP000007062 UP000235965 UP000076858
UP000008820 UP000192223 UP000053105 UP000069940 UP000249989 UP000008237 UP000075901 UP000075881 UP000007266 UP000002320 UP000075886 UP000075903 UP000075882 UP000076407 UP000075902 UP000075840 UP000007062 UP000235965 UP000076858
Pfam
PF03098 An_peroxidase
Interpro
SUPFAM
SSF48113
SSF48113
Gene 3D
ProteinModelPortal
H9JT75
A0A2W1BT77
A0A088MGW5
A0A2A4JI21
H9JT76
A0A2H1WGR9
+ More
A0A2H1W8U7 I4DS54 I4DN32 A0A194Q7Y0 I4DJ10 A0A194RBC8 A0A194Q9H2 A0A194RAP6 A0A194QDV6 A0A212ETD1 A0A1E1WUQ6 A0A2H1W9U4 A0A2W1BYE5 A0A2A4IUC0 H9ISL2 A0A2W1BT67 A0A3S2NLC3 A0A2A4J7D3 A0A2H1W4U4 A0A3S2LP82 A0A2H1WBG2 A0A2H1VUX0 A0A437AS20 A0A437ASV3 A0A2H1V5L5 O96645 Q177U0 Q16E76 A0A437ARC6 A0A1W4XPE3 W8C2M9 H9JQT0 A0A0P6H2D0 A0A0M9AC65 A0A182G7N2 A0A1B6I2F8 A0A1W6EVW7 E2BYM5 A0A1B6KS40 A0A0P6FAZ7 A0A1B6CGS3 A0A1B6FKB9 A0A0N8E1B0 A0A437AYB0 A0A1B6ME87 A0A1B6D0W8 A0A0P5RF89 A0A182T872 A0A182KBY5 A0A0N8EAV2 A0A0P6DND4 A0A0P6DE26 A0A0P6DDQ5 A0A0P6DGA0 A0A0P6GA01 A0A0P5PP69 A0A0P6FNT6 A0A0P5QCR3 A0A0P6E2J6 D6WB23 A0A0P6E456 A0A0P6F2H6 B0XE84 B0WM28 A0A139WNI1 A0A182Q218 A0A182VL61 A0A182KV49 A0A182WXU8 A0A1S4H4U5 A0A182TM70 A0A182I5W9 Q7QDZ3 A0A0K8VYA7 A0A0P4ZAS8 A0A0P6FX89 A0A0P4ZCC9 H9JT78 A0A0P5JNG9 A0A0P5SA74 A0A0P5M0T2 A0A0N8CDW0 A0A2J7R3R2 A0A0N8BFF0 A0A0P5SIR0 A0A0P6GB44 A0A0P6FJQ0 A0A0P6DU56 A0A164VLV1 A0A0P5VUV2 A0A0P5NKS6 A0A0P5WBR2 A0A0P5SKA9
A0A2H1W8U7 I4DS54 I4DN32 A0A194Q7Y0 I4DJ10 A0A194RBC8 A0A194Q9H2 A0A194RAP6 A0A194QDV6 A0A212ETD1 A0A1E1WUQ6 A0A2H1W9U4 A0A2W1BYE5 A0A2A4IUC0 H9ISL2 A0A2W1BT67 A0A3S2NLC3 A0A2A4J7D3 A0A2H1W4U4 A0A3S2LP82 A0A2H1WBG2 A0A2H1VUX0 A0A437AS20 A0A437ASV3 A0A2H1V5L5 O96645 Q177U0 Q16E76 A0A437ARC6 A0A1W4XPE3 W8C2M9 H9JQT0 A0A0P6H2D0 A0A0M9AC65 A0A182G7N2 A0A1B6I2F8 A0A1W6EVW7 E2BYM5 A0A1B6KS40 A0A0P6FAZ7 A0A1B6CGS3 A0A1B6FKB9 A0A0N8E1B0 A0A437AYB0 A0A1B6ME87 A0A1B6D0W8 A0A0P5RF89 A0A182T872 A0A182KBY5 A0A0N8EAV2 A0A0P6DND4 A0A0P6DE26 A0A0P6DDQ5 A0A0P6DGA0 A0A0P6GA01 A0A0P5PP69 A0A0P6FNT6 A0A0P5QCR3 A0A0P6E2J6 D6WB23 A0A0P6E456 A0A0P6F2H6 B0XE84 B0WM28 A0A139WNI1 A0A182Q218 A0A182VL61 A0A182KV49 A0A182WXU8 A0A1S4H4U5 A0A182TM70 A0A182I5W9 Q7QDZ3 A0A0K8VYA7 A0A0P4ZAS8 A0A0P6FX89 A0A0P4ZCC9 H9JT78 A0A0P5JNG9 A0A0P5SA74 A0A0P5M0T2 A0A0N8CDW0 A0A2J7R3R2 A0A0N8BFF0 A0A0P5SIR0 A0A0P6GB44 A0A0P6FJQ0 A0A0P6DU56 A0A164VLV1 A0A0P5VUV2 A0A0P5NKS6 A0A0P5WBR2 A0A0P5SKA9
PDB
3R5Q
E-value=5.77206e-12,
Score=166
Ontologies
GO
PANTHER
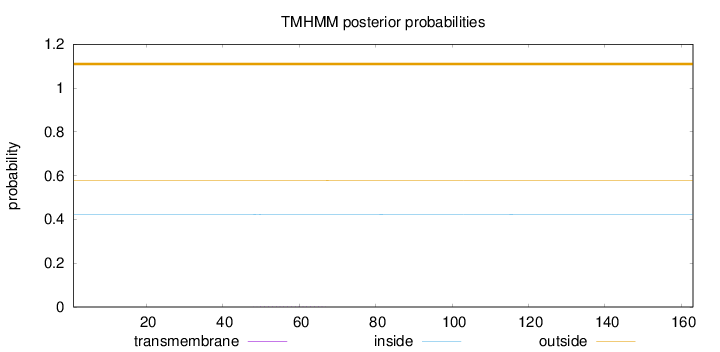
Topology
Length:
163
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02799
Exp number, first 60 AAs:
0.01634
Total prob of N-in:
0.42344
outside
1 - 163
Population Genetic Test Statistics
Pi
236.751142
Theta
203.703399
Tajima's D
1.594607
CLR
4.468915
CSRT
0.809259537023149
Interpretation
Uncertain
