Gene
KWMTBOMO12810 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012740
Annotation
PREDICTED:_peroxidase-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.702
Sequence
CDS
ATGCTCTGGTCATTAGTTTTCTTGGCGTTTCTGAAGCTGACCGCCGGTGTTCGCTATGAATCGTTTTCTGGTCTACCAGCGACTGATGAAAAGATCCAATACTTCATCAAACAGGAAAATTTAGATAACTGCACCAATGACATTCAACTCCCCTGCGACCCCAATGAGAGACGACGTCTAGATGGCTCATGCAGTAATTTCGACTATCCGTCAAGAGGAACATTCCATACCCCTGTGATAAGATTGTTACCCGAAGCCGACTGTGGAGACGATGAAACCTCTCCATCTGGAGCTCCTTTAAAATCTGCCCGGGAAGTCAGGCAGCGTATTTTACAGACAGGGAAGGCATCAGACCTCTCATACACACAACTATTGGCGATTATCTCGACAATCATATTTGCTGACCTTGGTTCAATCCATGATTCCGTGAATTTGTTAACGGAGACCACAAATTGTTGCACAGCGGAAGGAAAATCAAATTATATGTGCACGCCTATAGATATTCCCCAGGACGACCCTGTCCATAGGTTCTCTGGTATACGCTGCTTGAATCTCACTCGACCGAAATCGTTCCAGACTTATGGCTGCCTTGCTGACTCGAATGTAGACAGAATCGAATTCACAACACCGTTATTTGATTTATCGACTATCTACCGATCGACCGAACCGGCTCCAGAATACAGAACATACAGTGGCGGATTACTGATGACCCAAGAGGCCGATGGCACCATCTTCCCCCCACAAGAAGGACCGCACAGCAATAAATGCTTGCAAAACGACGCGTCTAATGGAGAAACGAAATGTTTTGGCCCTGTTTCAACATCGATTCTGCCGGTCACATTGTTGGTCGTGTGGTGGTGGAGACTACACAATAAGATCGCCAAAGAACTAAACGAAATCAATCCTCATTGGGATGACGAAACATTGTTCCAAACTGCAAGGGACATTAACATCGCAATAACAAACCAATTTGTTTACTACGAATTGCTGCCAACTCTATTCGGTGAAGAATTCTGTTTGAAAAATGAGCTGATTCACTCTGAATCTGGACACAGAGACCTTTACGATGAGTCTATACCTGCTACTTATCTCGAATACTACCTCGCTTTGAGATGGTTCCACTTGGTTTCAGAGGGTGATTTGAAACTTTTCGACGAGGATTTCAAGTACGTGGGCAAAAAAATGGTAACAGACCTCTCTTTACATACTGATTTCTTGATGAGAGATAAGAACTTGGCAAAAATGACACGAGGAACTTACTATCAGGCTGGAGGAGATAATGATAGAGCTGTAGACCCGGCCATTTGTGATAAGGGATTAGGAATAATGCAAAAAGCATCCGACCTTACAGCAGCGGATTTGAGGAAAAATCAGCTTTTCAAAATTCCACCTTACGTCGACTACGTAAAACTGTGCCATGATGTCGAAATTAAAACATGGAAAGACATGTTGAAATTCATTGACATGGATCGAATTGAAAGTCTGCAAGAAATTTACGAAACCCCTGGTGACGTGGAACTCCTGGCAGGAATCTGGATTGAAAGACCTATGGAGGGAGGTTACGTTCCTCCAACCGCAGCTTGTATCATAAACAAACAACTGTCTCTCACTATGAAAGCTGACAGACATTGGTATGAGAGATCGGACAGACCCTACGCTTTCAATGTCGCTCAGCTGGCGGAAATAAGAAAAGCAACAGTTGCAGGCTTCCTTTGCGAAGTTGGTGACGGTGTAGAGAGAATCCAACGGGAGGCACTTAAAAGAGTGACTGCTTCGAATCCATTGGTCAGCTGCCAAGAAATCCCGAGATGGAATCTCGCTGCTTGGAAAGAATCTAAGTGA
Protein
MLWSLVFLAFLKLTAGVRYESFSGLPATDEKIQYFIKQENLDNCTNDIQLPCDPNERRRLDGSCSNFDYPSRGTFHTPVIRLLPEADCGDDETSPSGAPLKSAREVRQRILQTGKASDLSYTQLLAIISTIIFADLGSIHDSVNLLTETTNCCTAEGKSNYMCTPIDIPQDDPVHRFSGIRCLNLTRPKSFQTYGCLADSNVDRIEFTTPLFDLSTIYRSTEPAPEYRTYSGGLLMTQEADGTIFPPQEGPHSNKCLQNDASNGETKCFGPVSTSILPVTLLVVWWWRLHNKIAKELNEINPHWDDETLFQTARDINIAITNQFVYYELLPTLFGEEFCLKNELIHSESGHRDLYDESIPATYLEYYLALRWFHLVSEGDLKLFDEDFKYVGKKMVTDLSLHTDFLMRDKNLAKMTRGTYYQAGGDNDRAVDPAICDKGLGIMQKASDLTAADLRKNQLFKIPPYVDYVKLCHDVEIKTWKDMLKFIDMDRIESLQEIYETPGDVELLAGIWIERPMEGGYVPPTAACIINKQLSLTMKADRHWYERSDRPYAFNVAQLAEIRKATVAGFLCEVGDGVERIQREALKRVTASNPLVSCQEIPRWNLAAWKESK
Summary
Uniprot
H9JT78
A0A194Q7Y0
A0A194Q9H2
A0A194QDV6
I4DJ10
A0A2W1BT67
+ More
A0A2A4J7D3 A0A212ETD1 A0A2H1W9U4 A0A2H1WBG2 A0A194RAP6 A0A2A4IUC0 A0A3S2NLC3 A0A2W1BYE5 A0A194RBC8 A0A2H1VUX0 H9JT77 A0A2W1BT77 A0A2A4JI21 A0A088MGW5 A0A2H1WAX3 A0A2H1W8U7 H9JQT0 A0A437AYB0 A0A2W1BQ93 A0A2W1BS42 A0A2A4JM77 A0A2A4IUP3 A0A2W1BXH1 A0A2A4IUD5 H9ISL1 A0A194Q3N0 A0A2A4JMB0 A0A437AS20 A0A437ASV3 A0A0N0PED8 A0A3S2NMV9 H9IT78 A0A3S2M0V1 A0A3S2L850 A0A232F2X4 A0A2A3EU99 A0A088AE09 K7IYZ3 W5JRW3 A0A088MG66 A0A2M4ABL8 A0A2M4AAY9 A0A1Y9G8K4 B0WM28 B0XE84 E2BCE2 W8C2M9 A0A346D9R6 Q177U0 A0A2J7RAM0 A0A2M4DQF3 Q16E76 A0A1W4WMG7 A0A310SND6 A0A3L8DTW7 A0A182G7N2 O96645 A0A182IZD9 D6WCM3 A0A3S2M8S7 A0A0L7R2K3 A0A1J1J076 A0A182KV49 A0A182WXU8 A0A1S4H4U5 A0A182TM70 A0A182I5W9 Q7QDZ3 A0A067RJX3 A0A1I8NQ77 A0A026W3J3 A0A1I8NQ80 A0A182FTE8 A0A182Q218 A0A182VL61 A0A437BBT0 A0A1A9W584 A0A0A1WQI6 A0A336MK45 A0A084V9X5 A0A1B6MDD3 A0A2M3ZE85 A0A034WLG7 A0A034WQE0 A0A182PRL2 A0A182JJH9 A0A182KBY5 A0A1Y9H2I0 I1T1H0 A0A182UUG4
A0A2A4J7D3 A0A212ETD1 A0A2H1W9U4 A0A2H1WBG2 A0A194RAP6 A0A2A4IUC0 A0A3S2NLC3 A0A2W1BYE5 A0A194RBC8 A0A2H1VUX0 H9JT77 A0A2W1BT77 A0A2A4JI21 A0A088MGW5 A0A2H1WAX3 A0A2H1W8U7 H9JQT0 A0A437AYB0 A0A2W1BQ93 A0A2W1BS42 A0A2A4JM77 A0A2A4IUP3 A0A2W1BXH1 A0A2A4IUD5 H9ISL1 A0A194Q3N0 A0A2A4JMB0 A0A437AS20 A0A437ASV3 A0A0N0PED8 A0A3S2NMV9 H9IT78 A0A3S2M0V1 A0A3S2L850 A0A232F2X4 A0A2A3EU99 A0A088AE09 K7IYZ3 W5JRW3 A0A088MG66 A0A2M4ABL8 A0A2M4AAY9 A0A1Y9G8K4 B0WM28 B0XE84 E2BCE2 W8C2M9 A0A346D9R6 Q177U0 A0A2J7RAM0 A0A2M4DQF3 Q16E76 A0A1W4WMG7 A0A310SND6 A0A3L8DTW7 A0A182G7N2 O96645 A0A182IZD9 D6WCM3 A0A3S2M8S7 A0A0L7R2K3 A0A1J1J076 A0A182KV49 A0A182WXU8 A0A1S4H4U5 A0A182TM70 A0A182I5W9 Q7QDZ3 A0A067RJX3 A0A1I8NQ77 A0A026W3J3 A0A1I8NQ80 A0A182FTE8 A0A182Q218 A0A182VL61 A0A437BBT0 A0A1A9W584 A0A0A1WQI6 A0A336MK45 A0A084V9X5 A0A1B6MDD3 A0A2M3ZE85 A0A034WLG7 A0A034WQE0 A0A182PRL2 A0A182JJH9 A0A182KBY5 A0A1Y9H2I0 I1T1H0 A0A182UUG4
Pubmed
EMBL
BABH01034746
BABH01034747
KQ459324
KPJ01647.1
KPJ01645.1
KPJ01646.1
+ More
AK401278 BAM17900.1 KZ149907 PZC78258.1 NWSH01002854 PCG67434.1 AGBW02012596 OWR44740.1 ODYU01007226 SOQ49823.1 ODYU01007479 SOQ50286.1 KQ460416 KPJ14923.1 NWSH01006580 PCG63355.1 RSAL01000046 RVE50539.1 PZC78257.1 KPJ14922.1 ODYU01004596 SOQ44641.1 BABH01034743 BABH01034744 PZC78259.1 NWSH01001320 PCG71707.1 KJ995811 AIN39494.1 ODYU01007325 SOQ50002.1 ODYU01007063 SOQ49501.1 BABH01028746 BABH01028747 BABH01028748 RSAL01000273 RVE43156.1 KZ149956 PZC76461.1 PZC76464.1 NWSH01001116 PCG72533.1 NWSH01006414 PCG63451.1 PZC76463.1 NWSH01006200 PCG63597.1 BABH01013455 KQ459564 KPI99614.1 PCG72532.1 RSAL01001640 RVE40744.1 RSAL01000844 RVE41114.1 KQ459837 KPJ19761.1 RSAL01000231 RVE43837.1 BABH01013458 BABH01013459 BABH01013460 RSAL01000083 RVE48485.1 RVE41111.1 NNAY01001189 OXU24803.1 KZ288187 PBC34719.1 ADMH02000540 ETN66033.1 KJ995807 AIN39490.1 GGFK01004863 MBW38184.1 GGFK01004599 MBW37920.1 DS231994 EDS30856.1 DS232811 EDS45917.1 GL447257 EFN86678.1 GAMC01010556 JAB95999.1 MH606240 AXM43807.1 CH477371 EAT42428.1 NEVH01006567 PNF37876.1 GGFL01015608 MBW79786.1 CH478998 EAT32533.1 KQ760645 OAD59676.1 QOIP01000004 RLU23752.1 JXUM01046680 KQ561486 KXJ78429.1 AF098717 AAC97504.1 KQ971317 EEZ99172.1 RSAL01000011 RVE53586.1 KQ414666 KOC65001.1 CVRI01000064 CRL05204.1 AAAB01008848 APCN01003535 EAA07042.4 KK852427 KDR24107.1 KK107453 EZA50655.1 AXCN02001762 RSAL01000094 RVE47835.1 GBXI01013522 JAD00770.1 UFQT01001180 SSX29359.1 ATLV01000829 KE523910 KFB34769.1 GEBQ01006041 JAT33936.1 GGFM01006040 MBW26791.1 GAKP01002541 JAC56411.1 GAKP01002543 GAKP01002542 JAC56410.1 JN003585 AEJ88361.1
AK401278 BAM17900.1 KZ149907 PZC78258.1 NWSH01002854 PCG67434.1 AGBW02012596 OWR44740.1 ODYU01007226 SOQ49823.1 ODYU01007479 SOQ50286.1 KQ460416 KPJ14923.1 NWSH01006580 PCG63355.1 RSAL01000046 RVE50539.1 PZC78257.1 KPJ14922.1 ODYU01004596 SOQ44641.1 BABH01034743 BABH01034744 PZC78259.1 NWSH01001320 PCG71707.1 KJ995811 AIN39494.1 ODYU01007325 SOQ50002.1 ODYU01007063 SOQ49501.1 BABH01028746 BABH01028747 BABH01028748 RSAL01000273 RVE43156.1 KZ149956 PZC76461.1 PZC76464.1 NWSH01001116 PCG72533.1 NWSH01006414 PCG63451.1 PZC76463.1 NWSH01006200 PCG63597.1 BABH01013455 KQ459564 KPI99614.1 PCG72532.1 RSAL01001640 RVE40744.1 RSAL01000844 RVE41114.1 KQ459837 KPJ19761.1 RSAL01000231 RVE43837.1 BABH01013458 BABH01013459 BABH01013460 RSAL01000083 RVE48485.1 RVE41111.1 NNAY01001189 OXU24803.1 KZ288187 PBC34719.1 ADMH02000540 ETN66033.1 KJ995807 AIN39490.1 GGFK01004863 MBW38184.1 GGFK01004599 MBW37920.1 DS231994 EDS30856.1 DS232811 EDS45917.1 GL447257 EFN86678.1 GAMC01010556 JAB95999.1 MH606240 AXM43807.1 CH477371 EAT42428.1 NEVH01006567 PNF37876.1 GGFL01015608 MBW79786.1 CH478998 EAT32533.1 KQ760645 OAD59676.1 QOIP01000004 RLU23752.1 JXUM01046680 KQ561486 KXJ78429.1 AF098717 AAC97504.1 KQ971317 EEZ99172.1 RSAL01000011 RVE53586.1 KQ414666 KOC65001.1 CVRI01000064 CRL05204.1 AAAB01008848 APCN01003535 EAA07042.4 KK852427 KDR24107.1 KK107453 EZA50655.1 AXCN02001762 RSAL01000094 RVE47835.1 GBXI01013522 JAD00770.1 UFQT01001180 SSX29359.1 ATLV01000829 KE523910 KFB34769.1 GEBQ01006041 JAT33936.1 GGFM01006040 MBW26791.1 GAKP01002541 JAC56411.1 GAKP01002543 GAKP01002542 JAC56410.1 JN003585 AEJ88361.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000053240
UP000283053
+ More
UP000215335 UP000242457 UP000005203 UP000002358 UP000000673 UP000069272 UP000002320 UP000008237 UP000008820 UP000235965 UP000192223 UP000279307 UP000069940 UP000249989 UP000075880 UP000007266 UP000053825 UP000183832 UP000075882 UP000076407 UP000075902 UP000075840 UP000007062 UP000027135 UP000095300 UP000053097 UP000075886 UP000075903 UP000091820 UP000030765 UP000075885 UP000075881 UP000075884
UP000215335 UP000242457 UP000005203 UP000002358 UP000000673 UP000069272 UP000002320 UP000008237 UP000008820 UP000235965 UP000192223 UP000279307 UP000069940 UP000249989 UP000075880 UP000007266 UP000053825 UP000183832 UP000075882 UP000076407 UP000075902 UP000075840 UP000007062 UP000027135 UP000095300 UP000053097 UP000075886 UP000075903 UP000091820 UP000030765 UP000075885 UP000075881 UP000075884
Pfam
Interpro
IPR010255
Haem_peroxidase_sf
+ More
IPR029585 Pxd-like
IPR019791 Haem_peroxidase_animal
IPR037120 Haem_peroxidase_sf_animal
IPR005828 MFS_sugar_transport-like
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
IPR012337 RNaseH-like_sf
IPR011641 Tyr-kin_ephrin_A/B_rcpt-like
IPR008979 Galactose-bd-like_sf
IPR001426 Tyr_kinase_rcpt_V_CS
IPR003961 FN3_dom
IPR027936 Eph_TM
IPR009030 Growth_fac_rcpt_cys_sf
IPR013783 Ig-like_fold
IPR013761 SAM/pointed_sf
IPR001090 Ephrin_rcpt_lig-bd_dom
IPR036116 FN3_sf
IPR001660 SAM
IPR029585 Pxd-like
IPR019791 Haem_peroxidase_animal
IPR037120 Haem_peroxidase_sf_animal
IPR005828 MFS_sugar_transport-like
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
IPR012337 RNaseH-like_sf
IPR011641 Tyr-kin_ephrin_A/B_rcpt-like
IPR008979 Galactose-bd-like_sf
IPR001426 Tyr_kinase_rcpt_V_CS
IPR003961 FN3_dom
IPR027936 Eph_TM
IPR009030 Growth_fac_rcpt_cys_sf
IPR013783 Ig-like_fold
IPR013761 SAM/pointed_sf
IPR001090 Ephrin_rcpt_lig-bd_dom
IPR036116 FN3_sf
IPR001660 SAM
SUPFAM
Gene 3D
ProteinModelPortal
H9JT78
A0A194Q7Y0
A0A194Q9H2
A0A194QDV6
I4DJ10
A0A2W1BT67
+ More
A0A2A4J7D3 A0A212ETD1 A0A2H1W9U4 A0A2H1WBG2 A0A194RAP6 A0A2A4IUC0 A0A3S2NLC3 A0A2W1BYE5 A0A194RBC8 A0A2H1VUX0 H9JT77 A0A2W1BT77 A0A2A4JI21 A0A088MGW5 A0A2H1WAX3 A0A2H1W8U7 H9JQT0 A0A437AYB0 A0A2W1BQ93 A0A2W1BS42 A0A2A4JM77 A0A2A4IUP3 A0A2W1BXH1 A0A2A4IUD5 H9ISL1 A0A194Q3N0 A0A2A4JMB0 A0A437AS20 A0A437ASV3 A0A0N0PED8 A0A3S2NMV9 H9IT78 A0A3S2M0V1 A0A3S2L850 A0A232F2X4 A0A2A3EU99 A0A088AE09 K7IYZ3 W5JRW3 A0A088MG66 A0A2M4ABL8 A0A2M4AAY9 A0A1Y9G8K4 B0WM28 B0XE84 E2BCE2 W8C2M9 A0A346D9R6 Q177U0 A0A2J7RAM0 A0A2M4DQF3 Q16E76 A0A1W4WMG7 A0A310SND6 A0A3L8DTW7 A0A182G7N2 O96645 A0A182IZD9 D6WCM3 A0A3S2M8S7 A0A0L7R2K3 A0A1J1J076 A0A182KV49 A0A182WXU8 A0A1S4H4U5 A0A182TM70 A0A182I5W9 Q7QDZ3 A0A067RJX3 A0A1I8NQ77 A0A026W3J3 A0A1I8NQ80 A0A182FTE8 A0A182Q218 A0A182VL61 A0A437BBT0 A0A1A9W584 A0A0A1WQI6 A0A336MK45 A0A084V9X5 A0A1B6MDD3 A0A2M3ZE85 A0A034WLG7 A0A034WQE0 A0A182PRL2 A0A182JJH9 A0A182KBY5 A0A1Y9H2I0 I1T1H0 A0A182UUG4
A0A2A4J7D3 A0A212ETD1 A0A2H1W9U4 A0A2H1WBG2 A0A194RAP6 A0A2A4IUC0 A0A3S2NLC3 A0A2W1BYE5 A0A194RBC8 A0A2H1VUX0 H9JT77 A0A2W1BT77 A0A2A4JI21 A0A088MGW5 A0A2H1WAX3 A0A2H1W8U7 H9JQT0 A0A437AYB0 A0A2W1BQ93 A0A2W1BS42 A0A2A4JM77 A0A2A4IUP3 A0A2W1BXH1 A0A2A4IUD5 H9ISL1 A0A194Q3N0 A0A2A4JMB0 A0A437AS20 A0A437ASV3 A0A0N0PED8 A0A3S2NMV9 H9IT78 A0A3S2M0V1 A0A3S2L850 A0A232F2X4 A0A2A3EU99 A0A088AE09 K7IYZ3 W5JRW3 A0A088MG66 A0A2M4ABL8 A0A2M4AAY9 A0A1Y9G8K4 B0WM28 B0XE84 E2BCE2 W8C2M9 A0A346D9R6 Q177U0 A0A2J7RAM0 A0A2M4DQF3 Q16E76 A0A1W4WMG7 A0A310SND6 A0A3L8DTW7 A0A182G7N2 O96645 A0A182IZD9 D6WCM3 A0A3S2M8S7 A0A0L7R2K3 A0A1J1J076 A0A182KV49 A0A182WXU8 A0A1S4H4U5 A0A182TM70 A0A182I5W9 Q7QDZ3 A0A067RJX3 A0A1I8NQ77 A0A026W3J3 A0A1I8NQ80 A0A182FTE8 A0A182Q218 A0A182VL61 A0A437BBT0 A0A1A9W584 A0A0A1WQI6 A0A336MK45 A0A084V9X5 A0A1B6MDD3 A0A2M3ZE85 A0A034WLG7 A0A034WQE0 A0A182PRL2 A0A182JJH9 A0A182KBY5 A0A1Y9H2I0 I1T1H0 A0A182UUG4
PDB
3R5Q
E-value=1.4427e-28,
Score=316
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.982412
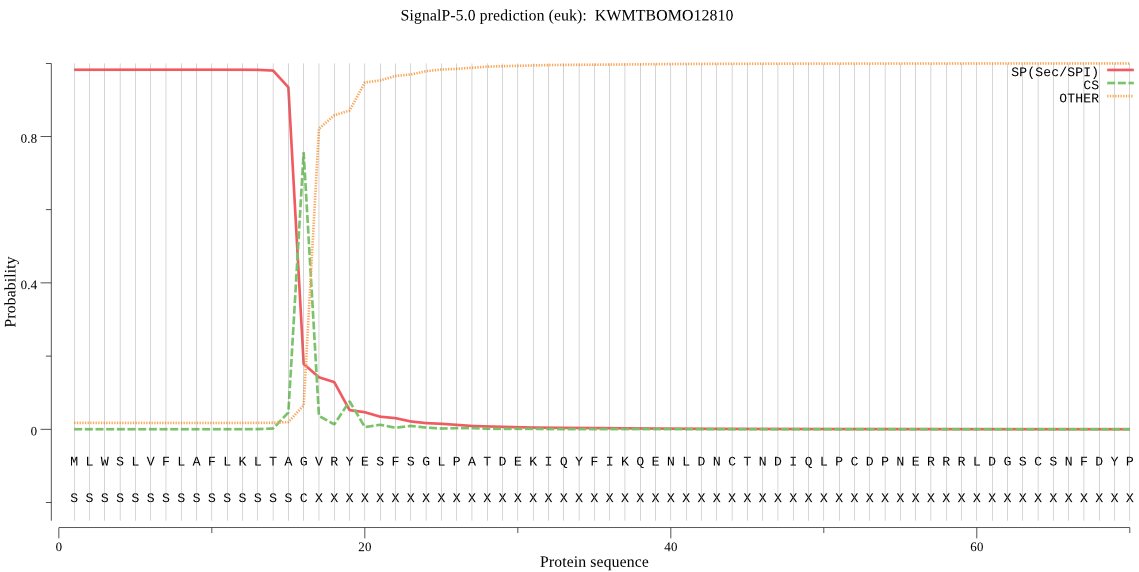
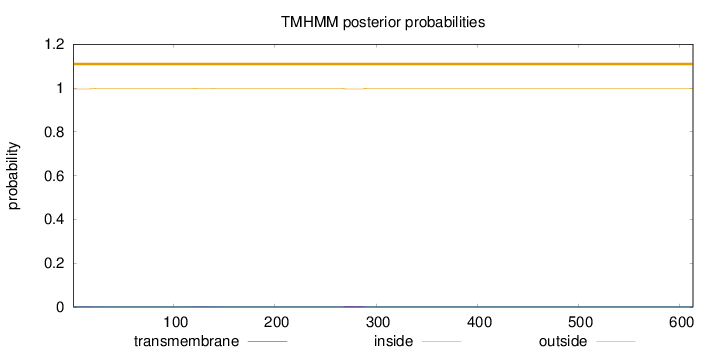
Length:
613
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17189
Exp number, first 60 AAs:
0.03971
Total prob of N-in:
0.00380
outside
1 - 613
Population Genetic Test Statistics
Pi
171.660697
Theta
126.845512
Tajima's D
0.945588
CLR
1.20935
CSRT
0.64046797660117
Interpretation
Uncertain
