Gene
KWMTBOMO12809
Pre Gene Modal
BGIBMGA012739
Annotation
PREDICTED:_peroxidase-like_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 2.841
Sequence
CDS
ATGGAAAGGCTGATAAAAGACGGGGTTATATCAAAACACAAAGGCTACAGAGATGTTTATGATGAGAAGATTATACCACAAATGTCCGACGAGTATTCTTATGTGCTACGATGGTTTCACATTGCACAAGAAGCGACCCTTGAATTGTACGATGAAAACTACAAGTGCTTCAAGACATTCCCTATGGTCAATTTGACCACCAGAACAGCATACTTCCCTGAAAATGACCATGAAGCCCAGATAGCTCGTGGTAGTTTCTTGCAGTCCACTTCTAAATTTATGGACCATGCTGTGGATCCTGATGTTGGTATTAAAATACGCGCATATGGCTTAGTTAAATAG
Protein
MERLIKDGVISKHKGYRDVYDEKIIPQMSDEYSYVLRWFHIAQEATLELYDENYKCFKTFPMVNLTTRTAYFPENDHEAQIARGSFLQSTSKFMDHAVDPDVGIKIRAYGLVK
Summary
Uniprot
H9JT77
A0A2H1WGR9
A0A194RAP6
A0A2W1BYE5
A0A194QDV6
I4DS54
+ More
A0A194RBC8 A0A2A4IUC0 A0A194Q9H2 I4DJ10 A0A2H1WBG2 A0A194Q7Y0 A0A3S2NLC3 A0A1E1WFW9 A0A2H1W4U4 A0A2H1W9U4 A0A2H1W8U7 A0A2H1WAX3 A0A088MGW5 A0A2W1BT67 H9ISL1 A0A2A4JI21 H9ISL2 A0A2W1BT77 A0A2A4J7D3 A0A437ASD3 A0A437AS20 A0A3S2L850 A0A212ETD1 A0A437ASV3 A0A3S2NMV9 H9JQT0 A0A194Q3N0 A0A2H1VUX0 A0A0N0PED8 H9JT78 H9IT78 A0A437BBT0 A0A2W1BQ93 A0A2A4IUP3 A0A437AYB0 A0A0L7LKV1 A0A2W1BXH1 A0A2W1BS42 A0A2A4JMB0 A0A2A4JM77 A0A2A4IUD5
A0A194RBC8 A0A2A4IUC0 A0A194Q9H2 I4DJ10 A0A2H1WBG2 A0A194Q7Y0 A0A3S2NLC3 A0A1E1WFW9 A0A2H1W4U4 A0A2H1W9U4 A0A2H1W8U7 A0A2H1WAX3 A0A088MGW5 A0A2W1BT67 H9ISL1 A0A2A4JI21 H9ISL2 A0A2W1BT77 A0A2A4J7D3 A0A437ASD3 A0A437AS20 A0A3S2L850 A0A212ETD1 A0A437ASV3 A0A3S2NMV9 H9JQT0 A0A194Q3N0 A0A2H1VUX0 A0A0N0PED8 H9JT78 H9IT78 A0A437BBT0 A0A2W1BQ93 A0A2A4IUP3 A0A437AYB0 A0A0L7LKV1 A0A2W1BXH1 A0A2W1BS42 A0A2A4JMB0 A0A2A4JM77 A0A2A4IUD5
EMBL
BABH01034743
BABH01034744
ODYU01008556
SOQ52260.1
KQ460416
KPJ14923.1
+ More
KZ149907 PZC78257.1 KQ459324 KPJ01646.1 AK405426 BAM20744.1 KPJ14922.1 NWSH01006580 PCG63355.1 KPJ01645.1 AK401278 BAM17900.1 ODYU01007479 SOQ50286.1 KPJ01647.1 RSAL01000046 RVE50539.1 GDQN01005160 JAT85894.1 ODYU01006337 SOQ48101.1 ODYU01007226 SOQ49823.1 ODYU01007063 SOQ49501.1 ODYU01007325 SOQ50002.1 KJ995811 AIN39494.1 PZC78258.1 BABH01013455 NWSH01001320 PCG71707.1 BABH01013447 BABH01013448 PZC78259.1 NWSH01002854 PCG67434.1 RSAL01001106 RVE40943.1 RSAL01001640 RVE40744.1 RSAL01000844 RVE41111.1 AGBW02012596 OWR44740.1 RVE41114.1 RSAL01000231 RVE43837.1 BABH01028746 BABH01028747 BABH01028748 KQ459564 KPI99614.1 ODYU01004596 SOQ44641.1 KQ459837 KPJ19761.1 BABH01034746 BABH01034747 BABH01013458 BABH01013459 BABH01013460 RSAL01000094 RVE47835.1 KZ149956 PZC76461.1 NWSH01006414 PCG63451.1 RSAL01000273 RVE43156.1 JTDY01000732 KOB76072.1 PZC76463.1 PZC76464.1 NWSH01001116 PCG72532.1 PCG72533.1 NWSH01006200 PCG63597.1
KZ149907 PZC78257.1 KQ459324 KPJ01646.1 AK405426 BAM20744.1 KPJ14922.1 NWSH01006580 PCG63355.1 KPJ01645.1 AK401278 BAM17900.1 ODYU01007479 SOQ50286.1 KPJ01647.1 RSAL01000046 RVE50539.1 GDQN01005160 JAT85894.1 ODYU01006337 SOQ48101.1 ODYU01007226 SOQ49823.1 ODYU01007063 SOQ49501.1 ODYU01007325 SOQ50002.1 KJ995811 AIN39494.1 PZC78258.1 BABH01013455 NWSH01001320 PCG71707.1 BABH01013447 BABH01013448 PZC78259.1 NWSH01002854 PCG67434.1 RSAL01001106 RVE40943.1 RSAL01001640 RVE40744.1 RSAL01000844 RVE41111.1 AGBW02012596 OWR44740.1 RVE41114.1 RSAL01000231 RVE43837.1 BABH01028746 BABH01028747 BABH01028748 KQ459564 KPI99614.1 ODYU01004596 SOQ44641.1 KQ459837 KPJ19761.1 BABH01034746 BABH01034747 BABH01013458 BABH01013459 BABH01013460 RSAL01000094 RVE47835.1 KZ149956 PZC76461.1 NWSH01006414 PCG63451.1 RSAL01000273 RVE43156.1 JTDY01000732 KOB76072.1 PZC76463.1 PZC76464.1 NWSH01001116 PCG72532.1 PCG72533.1 NWSH01006200 PCG63597.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JT77
A0A2H1WGR9
A0A194RAP6
A0A2W1BYE5
A0A194QDV6
I4DS54
+ More
A0A194RBC8 A0A2A4IUC0 A0A194Q9H2 I4DJ10 A0A2H1WBG2 A0A194Q7Y0 A0A3S2NLC3 A0A1E1WFW9 A0A2H1W4U4 A0A2H1W9U4 A0A2H1W8U7 A0A2H1WAX3 A0A088MGW5 A0A2W1BT67 H9ISL1 A0A2A4JI21 H9ISL2 A0A2W1BT77 A0A2A4J7D3 A0A437ASD3 A0A437AS20 A0A3S2L850 A0A212ETD1 A0A437ASV3 A0A3S2NMV9 H9JQT0 A0A194Q3N0 A0A2H1VUX0 A0A0N0PED8 H9JT78 H9IT78 A0A437BBT0 A0A2W1BQ93 A0A2A4IUP3 A0A437AYB0 A0A0L7LKV1 A0A2W1BXH1 A0A2W1BS42 A0A2A4JMB0 A0A2A4JM77 A0A2A4IUD5
A0A194RBC8 A0A2A4IUC0 A0A194Q9H2 I4DJ10 A0A2H1WBG2 A0A194Q7Y0 A0A3S2NLC3 A0A1E1WFW9 A0A2H1W4U4 A0A2H1W9U4 A0A2H1W8U7 A0A2H1WAX3 A0A088MGW5 A0A2W1BT67 H9ISL1 A0A2A4JI21 H9ISL2 A0A2W1BT77 A0A2A4J7D3 A0A437ASD3 A0A437AS20 A0A3S2L850 A0A212ETD1 A0A437ASV3 A0A3S2NMV9 H9JQT0 A0A194Q3N0 A0A2H1VUX0 A0A0N0PED8 H9JT78 H9IT78 A0A437BBT0 A0A2W1BQ93 A0A2A4IUP3 A0A437AYB0 A0A0L7LKV1 A0A2W1BXH1 A0A2W1BS42 A0A2A4JMB0 A0A2A4JM77 A0A2A4IUD5
Ontologies
PANTHER
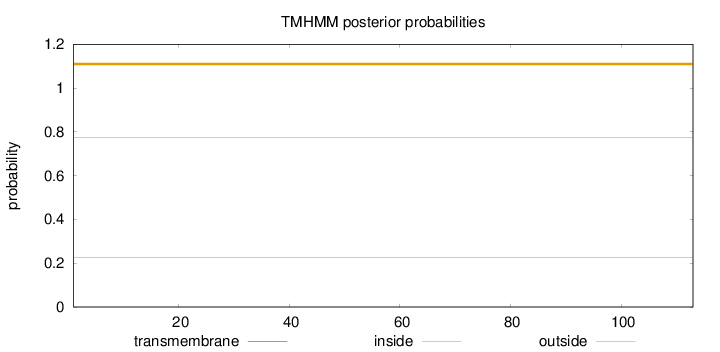
Topology
Length:
113
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.22747
outside
1 - 113
Population Genetic Test Statistics
Pi
260.540495
Theta
165.626022
Tajima's D
2.09159
CLR
0.212902
CSRT
0.896605169741513
Interpretation
Uncertain
