Gene
KWMTBOMO12796
Annotation
hypothetical_protein_M513_10814?_partial_[Trichuris_suis]
Location in the cell
Extracellular Reliability : 1.567 Nuclear Reliability : 1.877
Sequence
CDS
ATGGCCAGTCAAGAAATACAGGAACAACTGCAACAACTCTTGGCTGCAAATGCAGCGCTACAAAAACAACTTGAAGGAAATGAGAGAGAAACAAAATTCCTCCGACACCTACGCTCTCTTGCAGGAAGCACACTCACAGACGACAATATTATACGCCAACTTTGGTTACGCCGCTTGCCTCATCAGGCTCAAGCTATACTGGCAGCACAGGCTGACCTTGGCCTTGACAAATTGTCAGAACTAGCCGACAAAATTCTTGAGTTGGCCCAAACTCCTCCTATGGTATGTTCGACCGCAACATCTTCGAACAGTTTAACGCTTGGTTCATTAGCCAAACAACTAGAAGAGATCAGTAGCCAAGTTGCTGTCCTATCTAGGCGACAGAGCCGCCCTTATCGTGACAGATCATCATCTCGCGGCCCTAAAATTTGTTGATCACATTGA
Protein
MASQEIQEQLQQLLAANAALQKQLEGNERETKFLRHLRSLAGSTLTDDNIIRQLWLRRLPHQAQAILAAQADLGLDKLSELADKILELAQTPPMVCSTATSSNSLTLGSLAKQLEEISSQVAVLSRRQSRPYRDRSSSRGPKICXSH
Summary
Uniprot
A0A1E1W1E7
A0A085LTM6
A0A085N4U2
A0A085NBV3
A0A154PA69
A0A0A9Z2L8
+ More
A0A0J7KD10 T1HU96 A0A0J7K6Y3 A0A1W4X6B7 A0A0L7QK36 A0A0L7QJB5 A0A1B6JX45 A0A1W4XJL0 E9J0A8 A0A151IRN4 A0A2J7QH56 A0A131Y607 A0A183NPV5 A0A2J7RGP3 A0A151ITG2 A0A151IZ47 A0A151J3U8 A0A2J7PML8 X1XM89 A0A0J7K6I6 A0A094ZZD3 A0A151JSL7 S4PX01 A0A026X471 A0A085MBU7 A0A1W4XIW9 A0A151JCH0 A0A0L7QJ97 A0A183JUP6 A0A0J7K6V3 T1I831 A0A151J3D2 A0A026VRX8 A0A085LNY4 A0A085LIZ5 A0A151JUX4 A0A087UJA9 A0A131YTY1 A0A0L0ULM5 A0A2J7PD24 A0A2J7QJD3 A0A0J7K7T5 C4QPF1 A0A1Y1MXV9 A0A2L2YTR5 A0A151JWW0
A0A0J7KD10 T1HU96 A0A0J7K6Y3 A0A1W4X6B7 A0A0L7QK36 A0A0L7QJB5 A0A1B6JX45 A0A1W4XJL0 E9J0A8 A0A151IRN4 A0A2J7QH56 A0A131Y607 A0A183NPV5 A0A2J7RGP3 A0A151ITG2 A0A151IZ47 A0A151J3U8 A0A2J7PML8 X1XM89 A0A0J7K6I6 A0A094ZZD3 A0A151JSL7 S4PX01 A0A026X471 A0A085MBU7 A0A1W4XIW9 A0A151JCH0 A0A0L7QJ97 A0A183JUP6 A0A0J7K6V3 T1I831 A0A151J3D2 A0A026VRX8 A0A085LNY4 A0A085LIZ5 A0A151JUX4 A0A087UJA9 A0A131YTY1 A0A0L0ULM5 A0A2J7PD24 A0A2J7QJD3 A0A0J7K7T5 C4QPF1 A0A1Y1MXV9 A0A2L2YTR5 A0A151JWW0
Pubmed
EMBL
GDQN01010239
JAT80815.1
KL363297
KFD48322.1
KL367555
KFD64488.1
+ More
KL367519 KFD66949.1 KQ434840 KZC08080.1 GBHO01005513 JAG38091.1 LBMM01009142 KMQ88358.1 ACPB03016217 LBMM01012610 KMQ86112.1 KQ414987 KOC58988.1 LHQN01021927 LHQN01025379 KOC58628.1 KOC58659.1 GECU01003931 JAT03776.1 GL767374 EFZ13749.1 KQ981124 KYN09337.1 NEVH01014357 PNF27922.1 GEFM01002435 JAP73361.1 UZAL01010187 VDP03243.1 NEVH01003759 PNF40011.1 KQ981018 KYN10257.1 KQ980726 KYN14011.1 KQ980247 KYN17109.1 NEVH01023981 PNF17578.1 ABLF02055280 LBMM01013103 KMQ85816.1 KL251040 KGB38564.1 KQ978527 KYN30346.1 GAIX01005898 JAA86662.1 KK107029 EZA62169.1 KL363205 KFD54693.1 KQ979048 KYN23218.1 KQ415240 KOC58703.1 UZAK01014297 VDP04051.1 LBMM01012479 KMQ86203.1 ACPB03012735 KQ980296 KYN16851.1 KK111495 EZA46390.1 KL363358 KFD46680.1 KL364057 KL368168 KFD44941.1 KFD59316.1 KQ981772 KYN35826.1 KK120066 KFM77448.1 GEDV01006140 JAP82417.1 AJIL01003693 KNE87923.1 NEVH01026394 PNF14234.1 NEVH01013558 PNF28672.1 LBMM01012075 KMQ86422.1 CABG01000134 CAZ37338.2 GEZM01017686 JAV90494.1 IAAA01048245 LAA10725.1 KQ981619 KYN39245.1
KL367519 KFD66949.1 KQ434840 KZC08080.1 GBHO01005513 JAG38091.1 LBMM01009142 KMQ88358.1 ACPB03016217 LBMM01012610 KMQ86112.1 KQ414987 KOC58988.1 LHQN01021927 LHQN01025379 KOC58628.1 KOC58659.1 GECU01003931 JAT03776.1 GL767374 EFZ13749.1 KQ981124 KYN09337.1 NEVH01014357 PNF27922.1 GEFM01002435 JAP73361.1 UZAL01010187 VDP03243.1 NEVH01003759 PNF40011.1 KQ981018 KYN10257.1 KQ980726 KYN14011.1 KQ980247 KYN17109.1 NEVH01023981 PNF17578.1 ABLF02055280 LBMM01013103 KMQ85816.1 KL251040 KGB38564.1 KQ978527 KYN30346.1 GAIX01005898 JAA86662.1 KK107029 EZA62169.1 KL363205 KFD54693.1 KQ979048 KYN23218.1 KQ415240 KOC58703.1 UZAK01014297 VDP04051.1 LBMM01012479 KMQ86203.1 ACPB03012735 KQ980296 KYN16851.1 KK111495 EZA46390.1 KL363358 KFD46680.1 KL364057 KL368168 KFD44941.1 KFD59316.1 KQ981772 KYN35826.1 KK120066 KFM77448.1 GEDV01006140 JAP82417.1 AJIL01003693 KNE87923.1 NEVH01026394 PNF14234.1 NEVH01013558 PNF28672.1 LBMM01012075 KMQ86422.1 CABG01000134 CAZ37338.2 GEZM01017686 JAV90494.1 IAAA01048245 LAA10725.1 KQ981619 KYN39245.1
Proteomes
SUPFAM
SSF50630
SSF50630
Gene 3D
ProteinModelPortal
A0A1E1W1E7
A0A085LTM6
A0A085N4U2
A0A085NBV3
A0A154PA69
A0A0A9Z2L8
+ More
A0A0J7KD10 T1HU96 A0A0J7K6Y3 A0A1W4X6B7 A0A0L7QK36 A0A0L7QJB5 A0A1B6JX45 A0A1W4XJL0 E9J0A8 A0A151IRN4 A0A2J7QH56 A0A131Y607 A0A183NPV5 A0A2J7RGP3 A0A151ITG2 A0A151IZ47 A0A151J3U8 A0A2J7PML8 X1XM89 A0A0J7K6I6 A0A094ZZD3 A0A151JSL7 S4PX01 A0A026X471 A0A085MBU7 A0A1W4XIW9 A0A151JCH0 A0A0L7QJ97 A0A183JUP6 A0A0J7K6V3 T1I831 A0A151J3D2 A0A026VRX8 A0A085LNY4 A0A085LIZ5 A0A151JUX4 A0A087UJA9 A0A131YTY1 A0A0L0ULM5 A0A2J7PD24 A0A2J7QJD3 A0A0J7K7T5 C4QPF1 A0A1Y1MXV9 A0A2L2YTR5 A0A151JWW0
A0A0J7KD10 T1HU96 A0A0J7K6Y3 A0A1W4X6B7 A0A0L7QK36 A0A0L7QJB5 A0A1B6JX45 A0A1W4XJL0 E9J0A8 A0A151IRN4 A0A2J7QH56 A0A131Y607 A0A183NPV5 A0A2J7RGP3 A0A151ITG2 A0A151IZ47 A0A151J3U8 A0A2J7PML8 X1XM89 A0A0J7K6I6 A0A094ZZD3 A0A151JSL7 S4PX01 A0A026X471 A0A085MBU7 A0A1W4XIW9 A0A151JCH0 A0A0L7QJ97 A0A183JUP6 A0A0J7K6V3 T1I831 A0A151J3D2 A0A026VRX8 A0A085LNY4 A0A085LIZ5 A0A151JUX4 A0A087UJA9 A0A131YTY1 A0A0L0ULM5 A0A2J7PD24 A0A2J7QJD3 A0A0J7K7T5 C4QPF1 A0A1Y1MXV9 A0A2L2YTR5 A0A151JWW0
Ontologies
KEGG
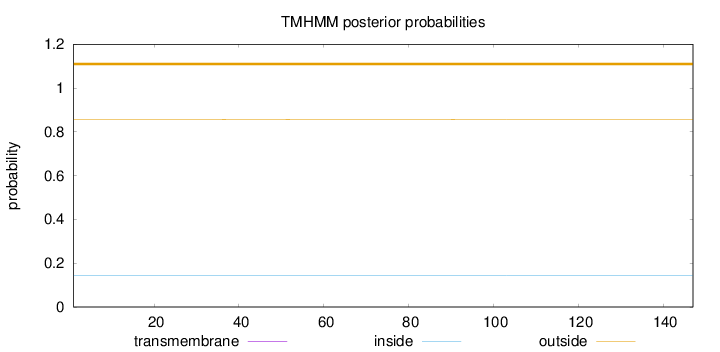
Topology
Length:
147
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00042
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.14333
outside
1 - 147
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
60.328768
CSRT
0
Interpretation
Uncertain
