Gene
KWMTBOMO12795
Annotation
PREDICTED:_cytochrome_P450_6k1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.251
Sequence
CDS
ATGGTTGCTGAACGGAGTAAAAGCCAAAGCATTTCGGGAAAGAGTAAAGATCTACTTGATGCTCTGATGGCCATGAAAATAGAGGCAGCTGCAGAGAATGAAGTTTACAACGAAGACCTATTATTTGCGCAAGCAACATTGTTTGTTCAAGCTGGATTTGAAACGACCTCGTCAGCAATTACTTTTGCTATTTATGAATTAGCTTACAATCCAGAAATACAGGAACGTCTGTACAGAGAGATAGTGGAAGCCAAACAGAAGATGGAGGGCAATGAACTTGATGGAGTAGTACTATCTAACTTACAATATCTTAACTGTGTTATTAATGAAACCCTACGGAAATACCCGTCTCTCGGTTGGTTAGACCGGGTATCTTCACAAAGCTACAAAGTGGACGATACGCTGACCGTACCAGCTGGAACTGCGGTGTATGTCAACGTGGCTGGGATCCAATCCGATCCTCAGCTATTTCCAAAACCTGAAGAGTTTATCCCGGAGCGGTTCAACACAGACAATAACAATATCAAGCCGTTTACTTTCATACCTTTTGGAGAAGGTCCTAGACAATGTATAGGTATACGTTTTGGCTATCAAGCGATACAATTTGGATTATCTGCAATTATTCTGAACTTTAAATTGCGTCCCATAGAAGGGTCACCGCTGCCTAACAACTGTCATATTGAAAGCAAAGGGTTTGTTTACACTGCCGATCATCCCTTGCATATACAGTTCGTGCCCAGGAACTGA
Protein
MVAERSKSQSISGKSKDLLDALMAMKIEAAAENEVYNEDLLFAQATLFVQAGFETTSSAITFAIYELAYNPEIQERLYREIVEAKQKMEGNELDGVVLSNLQYLNCVINETLRKYPSLGWLDRVSSQSYKVDDTLTVPAGTAVYVNVAGIQSDPQLFPKPEEFIPERFNTDNNNIKPFTFIPFGEGPRQCIGIRFGYQAIQFGLSAIILNFKLRPIEGSPLPNNCHIESKGFVYTADHPLHIQFVPRN
Summary
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Uniprot
A0A2H1V767
A0A2H1V738
A0A2W1BF80
A0A068EU25
A0A2W1BMN3
A0A194RM59
+ More
A0A3S2LYD4 A0A2A4J3Z1 A0A2W1BCF8 A0A286QUF5 A0A194QLW1 A0A1V0D9G1 A0A248QJ37 J7FJ25 A0A3S2NRQ2 A0A2A4JVD4 A0A0K8TV62 A0A2Z5CV07 A0A0C5C563 A0A1V0D9G8 A0A212ESQ5 A0A194QG26 A0A194RMF7 A0A212ET38 A0A2I6RJN8 A0A212FBK5 A0A1V0D9G2 A0A194QFW1 A0A2H1VSD4 J7FJ29 A0A1V0D9G0 A0A0L7LSI1 A0A194QHM5 A0A194QG95 A0A3G5EAS2 A0A286MXN6 A0A1V0D9H1 A0A2H1VRM6 A0A194QLW6 A0A1W4W447 A0A0T6B6B5 A0A194RR50 A0A2Z6JNJ2 D7EJT0 A0A0K1YWI1 A0A0K0LBA5 A0A194RE58 A0A385KZS5 A0A0T6AVG4 A0A1B6FE27 A0A1W6L1S3 A0A2P8XD24 A0A2Z6JQP2 A0A411K6Z1 A0A2Z6JSP9 A0A2S1PSN1 R9UDV4 A0A068F0U3 K4JN79 A0A154PSU2 A0A2P8Z5Z9 A0A2S1PSQ3 A0A1B6LYY8 D7EJT1 A0A1W6R7G7 E2AAV1 A0A2Z6JQN4 D5L0N2 A0A154PQK8 E0W389 A0A0T6AX95 A0A2Z6JNK9 A0A0T6BAW8 A0A194PQ11 A0A2Z5CVH9 A0A348G633 A0A182HQ31 A0A067R372 A0A1B6GJT5 A0A182KSN7 A0A0U4HU05 A0A1S4H8W2 D6WB59 A0A182XUH3 A0A182VDP4 A0A2Z6JNK2 A0A182TG63 A0A2Z6JJ49 A0A2Z6JJT5 A0A1L8E412 A0A2J7PXM6 A0A1L8E4K4 A0A2Z6JIS3 A0A2Z6JIS1 A0A067QX98 B6E554 E9IRU5 A0A1B0CD20 A0A0L0C9H1 A0A2J7QKF6
A0A3S2LYD4 A0A2A4J3Z1 A0A2W1BCF8 A0A286QUF5 A0A194QLW1 A0A1V0D9G1 A0A248QJ37 J7FJ25 A0A3S2NRQ2 A0A2A4JVD4 A0A0K8TV62 A0A2Z5CV07 A0A0C5C563 A0A1V0D9G8 A0A212ESQ5 A0A194QG26 A0A194RMF7 A0A212ET38 A0A2I6RJN8 A0A212FBK5 A0A1V0D9G2 A0A194QFW1 A0A2H1VSD4 J7FJ29 A0A1V0D9G0 A0A0L7LSI1 A0A194QHM5 A0A194QG95 A0A3G5EAS2 A0A286MXN6 A0A1V0D9H1 A0A2H1VRM6 A0A194QLW6 A0A1W4W447 A0A0T6B6B5 A0A194RR50 A0A2Z6JNJ2 D7EJT0 A0A0K1YWI1 A0A0K0LBA5 A0A194RE58 A0A385KZS5 A0A0T6AVG4 A0A1B6FE27 A0A1W6L1S3 A0A2P8XD24 A0A2Z6JQP2 A0A411K6Z1 A0A2Z6JSP9 A0A2S1PSN1 R9UDV4 A0A068F0U3 K4JN79 A0A154PSU2 A0A2P8Z5Z9 A0A2S1PSQ3 A0A1B6LYY8 D7EJT1 A0A1W6R7G7 E2AAV1 A0A2Z6JQN4 D5L0N2 A0A154PQK8 E0W389 A0A0T6AX95 A0A2Z6JNK9 A0A0T6BAW8 A0A194PQ11 A0A2Z5CVH9 A0A348G633 A0A182HQ31 A0A067R372 A0A1B6GJT5 A0A182KSN7 A0A0U4HU05 A0A1S4H8W2 D6WB59 A0A182XUH3 A0A182VDP4 A0A2Z6JNK2 A0A182TG63 A0A2Z6JJ49 A0A2Z6JJT5 A0A1L8E412 A0A2J7PXM6 A0A1L8E4K4 A0A2Z6JIS3 A0A2Z6JIS1 A0A067QX98 B6E554 E9IRU5 A0A1B0CD20 A0A0L0C9H1 A0A2J7QKF6
Pubmed
EMBL
ODYU01001012
SOQ36636.1
SOQ36637.1
KZ150141
PZC72941.1
KM016707
+ More
AID54859.1 PZC72943.1 KQ459995 KPJ18519.1 RSAL01000117 RVE46813.1 NWSH01003205 PCG66785.1 PZC72942.1 KX425013 ASN63840.1 KQ458981 KPJ04436.1 KY212076 ARA91640.1 KX443446 ASO98021.1 JX310086 AFP20597.1 RVE46814.1 NWSH01000502 PCG75997.1 GCVX01000057 JAI18173.1 MG583760 AXB26390.1 KP001135 AJN91180.1 KY212073 ARA91637.1 AGBW02012725 OWR44536.1 KPJ04442.1 KPJ18515.1 AGBW02012675 OWR44614.1 KY304478 AUN86409.1 AGBW02009303 OWR51116.1 KY212075 ARA91639.1 KPJ04438.1 ODYU01004176 SOQ43763.1 JX310101 AFP20612.1 KY212074 ARA91638.1 JTDY01000175 KOB78440.1 KPJ04440.1 KPJ04439.1 MH001164 AYW35337.1 MF684354 ASX93988.1 KY212072 ARA91636.1 ODYU01004035 SOQ43503.1 KPJ04441.1 LJIG01009517 KRT82922.1 KPJ18516.1 BK010372 DAB41775.1 DS497716 EFA12856.1 KP859393 AKZ17703.1 KM217024 AIW79987.1 KQ460367 KPJ15565.1 MG988388 AYA00974.1 LJIG01022791 KRT78695.1 GECZ01021328 JAS48441.1 KX609539 ARN17946.1 PYGN01002859 PSN29892.1 BK010508 DAB41791.1 MH500625 QBC73100.1 BK010376 DAB41779.1 MF278023 AWH61658.1 JX463007 AGN52800.1 KM016708 AID54860.1 JX876501 AFU86431.1 KQ435041 KZC14180.1 PYGN01000181 PSN51919.1 MF278024 AWH61659.1 GEBQ01024434 GEBQ01011137 JAT15543.1 JAT28840.1 EFA12857.1 KX687993 ARO50425.1 GL438178 EFN69444.1 BK010377 DAB41780.1 GU731537 ADE05587.1 KZC14179.1 DS235882 EEB20095.1 LJIG01022691 KRT79223.1 BK010513 DAB41796.1 LJIG01002452 KRT84488.1 KQ459602 KPI93220.1 MG583771 AXB26401.1 FX985572 BBF97906.1 APCN01003290 KK852786 KDR16543.1 GECZ01016884 GECZ01007084 JAS52885.1 JAS62685.1 KT071006 ALX81391.1 AAAB01008986 KQ971311 EEZ99338.1 BK010503 DAB41786.1 BK010378 DAB41781.1 BK010537 DAB41820.1 GFDF01000623 JAV13461.1 NEVH01020853 PNF21099.1 GFDF01000622 JAV13462.1 BK010502 DAB41785.1 BK010371 DAB41774.1 KK852906 KDR14000.1 FJ209361 ACI25098.1 GL765283 EFZ16666.1 AJWK01007320 AJWK01007321 JRES01000735 KNC28887.1 NEVH01013262 PNF29066.1
AID54859.1 PZC72943.1 KQ459995 KPJ18519.1 RSAL01000117 RVE46813.1 NWSH01003205 PCG66785.1 PZC72942.1 KX425013 ASN63840.1 KQ458981 KPJ04436.1 KY212076 ARA91640.1 KX443446 ASO98021.1 JX310086 AFP20597.1 RVE46814.1 NWSH01000502 PCG75997.1 GCVX01000057 JAI18173.1 MG583760 AXB26390.1 KP001135 AJN91180.1 KY212073 ARA91637.1 AGBW02012725 OWR44536.1 KPJ04442.1 KPJ18515.1 AGBW02012675 OWR44614.1 KY304478 AUN86409.1 AGBW02009303 OWR51116.1 KY212075 ARA91639.1 KPJ04438.1 ODYU01004176 SOQ43763.1 JX310101 AFP20612.1 KY212074 ARA91638.1 JTDY01000175 KOB78440.1 KPJ04440.1 KPJ04439.1 MH001164 AYW35337.1 MF684354 ASX93988.1 KY212072 ARA91636.1 ODYU01004035 SOQ43503.1 KPJ04441.1 LJIG01009517 KRT82922.1 KPJ18516.1 BK010372 DAB41775.1 DS497716 EFA12856.1 KP859393 AKZ17703.1 KM217024 AIW79987.1 KQ460367 KPJ15565.1 MG988388 AYA00974.1 LJIG01022791 KRT78695.1 GECZ01021328 JAS48441.1 KX609539 ARN17946.1 PYGN01002859 PSN29892.1 BK010508 DAB41791.1 MH500625 QBC73100.1 BK010376 DAB41779.1 MF278023 AWH61658.1 JX463007 AGN52800.1 KM016708 AID54860.1 JX876501 AFU86431.1 KQ435041 KZC14180.1 PYGN01000181 PSN51919.1 MF278024 AWH61659.1 GEBQ01024434 GEBQ01011137 JAT15543.1 JAT28840.1 EFA12857.1 KX687993 ARO50425.1 GL438178 EFN69444.1 BK010377 DAB41780.1 GU731537 ADE05587.1 KZC14179.1 DS235882 EEB20095.1 LJIG01022691 KRT79223.1 BK010513 DAB41796.1 LJIG01002452 KRT84488.1 KQ459602 KPI93220.1 MG583771 AXB26401.1 FX985572 BBF97906.1 APCN01003290 KK852786 KDR16543.1 GECZ01016884 GECZ01007084 JAS52885.1 JAS62685.1 KT071006 ALX81391.1 AAAB01008986 KQ971311 EEZ99338.1 BK010503 DAB41786.1 BK010378 DAB41781.1 BK010537 DAB41820.1 GFDF01000623 JAV13461.1 NEVH01020853 PNF21099.1 GFDF01000622 JAV13462.1 BK010502 DAB41785.1 BK010371 DAB41774.1 KK852906 KDR14000.1 FJ209361 ACI25098.1 GL765283 EFZ16666.1 AJWK01007320 AJWK01007321 JRES01000735 KNC28887.1 NEVH01013262 PNF29066.1
Proteomes
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
A0A2H1V767
A0A2H1V738
A0A2W1BF80
A0A068EU25
A0A2W1BMN3
A0A194RM59
+ More
A0A3S2LYD4 A0A2A4J3Z1 A0A2W1BCF8 A0A286QUF5 A0A194QLW1 A0A1V0D9G1 A0A248QJ37 J7FJ25 A0A3S2NRQ2 A0A2A4JVD4 A0A0K8TV62 A0A2Z5CV07 A0A0C5C563 A0A1V0D9G8 A0A212ESQ5 A0A194QG26 A0A194RMF7 A0A212ET38 A0A2I6RJN8 A0A212FBK5 A0A1V0D9G2 A0A194QFW1 A0A2H1VSD4 J7FJ29 A0A1V0D9G0 A0A0L7LSI1 A0A194QHM5 A0A194QG95 A0A3G5EAS2 A0A286MXN6 A0A1V0D9H1 A0A2H1VRM6 A0A194QLW6 A0A1W4W447 A0A0T6B6B5 A0A194RR50 A0A2Z6JNJ2 D7EJT0 A0A0K1YWI1 A0A0K0LBA5 A0A194RE58 A0A385KZS5 A0A0T6AVG4 A0A1B6FE27 A0A1W6L1S3 A0A2P8XD24 A0A2Z6JQP2 A0A411K6Z1 A0A2Z6JSP9 A0A2S1PSN1 R9UDV4 A0A068F0U3 K4JN79 A0A154PSU2 A0A2P8Z5Z9 A0A2S1PSQ3 A0A1B6LYY8 D7EJT1 A0A1W6R7G7 E2AAV1 A0A2Z6JQN4 D5L0N2 A0A154PQK8 E0W389 A0A0T6AX95 A0A2Z6JNK9 A0A0T6BAW8 A0A194PQ11 A0A2Z5CVH9 A0A348G633 A0A182HQ31 A0A067R372 A0A1B6GJT5 A0A182KSN7 A0A0U4HU05 A0A1S4H8W2 D6WB59 A0A182XUH3 A0A182VDP4 A0A2Z6JNK2 A0A182TG63 A0A2Z6JJ49 A0A2Z6JJT5 A0A1L8E412 A0A2J7PXM6 A0A1L8E4K4 A0A2Z6JIS3 A0A2Z6JIS1 A0A067QX98 B6E554 E9IRU5 A0A1B0CD20 A0A0L0C9H1 A0A2J7QKF6
A0A3S2LYD4 A0A2A4J3Z1 A0A2W1BCF8 A0A286QUF5 A0A194QLW1 A0A1V0D9G1 A0A248QJ37 J7FJ25 A0A3S2NRQ2 A0A2A4JVD4 A0A0K8TV62 A0A2Z5CV07 A0A0C5C563 A0A1V0D9G8 A0A212ESQ5 A0A194QG26 A0A194RMF7 A0A212ET38 A0A2I6RJN8 A0A212FBK5 A0A1V0D9G2 A0A194QFW1 A0A2H1VSD4 J7FJ29 A0A1V0D9G0 A0A0L7LSI1 A0A194QHM5 A0A194QG95 A0A3G5EAS2 A0A286MXN6 A0A1V0D9H1 A0A2H1VRM6 A0A194QLW6 A0A1W4W447 A0A0T6B6B5 A0A194RR50 A0A2Z6JNJ2 D7EJT0 A0A0K1YWI1 A0A0K0LBA5 A0A194RE58 A0A385KZS5 A0A0T6AVG4 A0A1B6FE27 A0A1W6L1S3 A0A2P8XD24 A0A2Z6JQP2 A0A411K6Z1 A0A2Z6JSP9 A0A2S1PSN1 R9UDV4 A0A068F0U3 K4JN79 A0A154PSU2 A0A2P8Z5Z9 A0A2S1PSQ3 A0A1B6LYY8 D7EJT1 A0A1W6R7G7 E2AAV1 A0A2Z6JQN4 D5L0N2 A0A154PQK8 E0W389 A0A0T6AX95 A0A2Z6JNK9 A0A0T6BAW8 A0A194PQ11 A0A2Z5CVH9 A0A348G633 A0A182HQ31 A0A067R372 A0A1B6GJT5 A0A182KSN7 A0A0U4HU05 A0A1S4H8W2 D6WB59 A0A182XUH3 A0A182VDP4 A0A2Z6JNK2 A0A182TG63 A0A2Z6JJ49 A0A2Z6JJT5 A0A1L8E412 A0A2J7PXM6 A0A1L8E4K4 A0A2Z6JIS3 A0A2Z6JIS1 A0A067QX98 B6E554 E9IRU5 A0A1B0CD20 A0A0L0C9H1 A0A2J7QKF6
PDB
6MJM
E-value=1.58082e-34,
Score=363
Ontologies
GO
Topology
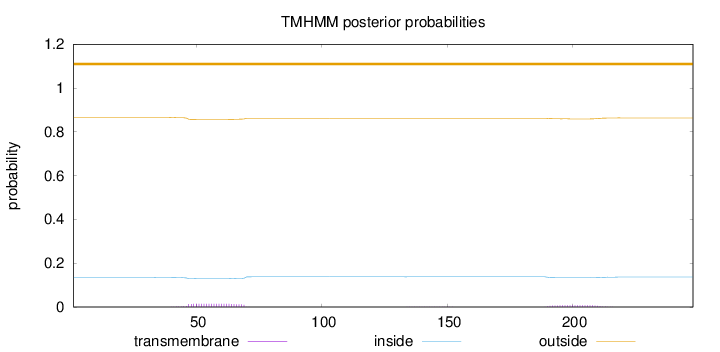
Length:
248
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.48785
Exp number, first 60 AAs:
0.2141
Total prob of N-in:
0.13509
outside
1 - 248
Population Genetic Test Statistics
Pi
14.265474
Theta
13.426115
Tajima's D
0.038171
CLR
0.470731
CSRT
0.388480575971201
Interpretation
Uncertain
