Pre Gene Modal
BGIBMGA007210
Annotation
PREDICTED:_ATP-dependent_Clp_protease_ATP-binding_subunit_clpX-like?_mitochondrial_isoform_X3_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.236 Nuclear Reliability : 1.774
Sequence
CDS
ATGGCCTTACCGGTGGCAAAGCGGAAACCTCCACCTCCGCCAAAGAAGATCTTCGAATATCTGAACAAGCACGTGGTGGGGCAGGAATACGCCAAGAAGGTGCTCTCGGTGGCCGTGTACAATCACTACAAACGCATCTACAACAACGTGACGAGCGGAACTCCGAACGACTCCCATCACCCGCTGCATCACACTCACAGAGATCTGCTACATTTGGGTCACGGAGGCAGCGGCGGTGGAGCGGAAGTATTGGAAAGGCAACACCATGACATCAAACTGGACAAAAGCAACGTACTACTGTTGGGACCCACCGGATGTGGTAAGACGCTGTTAGCACAAACGATAGCCCAGTGCCTGGACGTGCCGTTCGCGATCTGCGACTGCACCACGCTGACGCAGGCCGGCTACGTCGGCGAGGACATCGAGAGTGTCATCGCCAAACTGCTGCAGGACGCTAACTTCAACGTGGAGCGAGCTCAAACGGGGATAGTGTTCCTGGACGAGGTCGACAAAATCGGTGCCGTCCCGGGAATACATCAGCTCAGGGATGTCGGAGGCGAGGGAGTGCAGCAAGGCATGCTGAAGATGCTGGAGGGCGCCGTGGTGTCGGTGCCGGAGAGGAACTCTCGCAAGCTGCGTGGCGATGCGCTCAACGTCGACACCACCAACATCCTGTTCGTGGCCAGCGGCGCCTACAACGGGCTGGACAGGCTGGTGCAGCGGCGCAACAACGAGAAGTACCTGGGGTTCGGCGCGGGCGGCGCGGGCGGCGGCGCGGGGCGGCGGGCGGCGCTGCAGGCCGCGCACAACGACGCCAGCCCGGCCGCCGCGCCGCAGGACGAGGACCGCGAGCGCGACGCCTGGCTGCGCAAGGTGCAGGCCAGGGACCTCATCGACTTCGGCATGATACCGGAGTTTGTCGGGCGCTTCCCCGTGCTCGTGCCCTTCCACAGCCTCAGCCAGGACCTGCTGGTGCGGATCCTGACAGAACCTAAAAACGCGATGGTGGCGCAGTACAAGCTGCTGTTCGCGATGGACAAGTGCGAACTGTCGTTCAGCGACGACGCGCTGCGGGCCGTCGCCGCGCTCGCCATGGAGCGACGCACCGGCGCGCGCGGCCTGCGGGCCATCATGGAAAGTCTTCTGCTGGAGATAATGTTCGAGATCCCGGGGTCGGAGATATCCAGCGTGCACGTGCACGAGGGCTGCGTGTCCCGCGGCGAGCCCCCCCGCACCGCGCGCCGCGCCGCCGCCCCCGCGCCCGCCCCGCGCGACCCGCACCTGCACGACTGGCCCGAGCTCAGGATCACCAACTGTACGTACTGTTACAACCCTAGTGTGAAGAGCCCTAACCAAGCTGCACGATCAGCGTTCCCGAGCCCTCCTGTTCCGTCCGCTCAGCCAACATAA
Protein
MALPVAKRKPPPPPKKIFEYLNKHVVGQEYAKKVLSVAVYNHYKRIYNNVTSGTPNDSHHPLHHTHRDLLHLGHGGSGGGAEVLERQHHDIKLDKSNVLLLGPTGCGKTLLAQTIAQCLDVPFAICDCTTLTQAGYVGEDIESVIAKLLQDANFNVERAQTGIVFLDEVDKIGAVPGIHQLRDVGGEGVQQGMLKMLEGAVVSVPERNSRKLRGDALNVDTTNILFVASGAYNGLDRLVQRRNNEKYLGFGAGGAGGGAGRRAALQAAHNDASPAAAPQDEDRERDAWLRKVQARDLIDFGMIPEFVGRFPVLVPFHSLSQDLLVRILTEPKNAMVAQYKLLFAMDKCELSFSDDALRAVAALAMERRTGARGLRAIMESLLLEIMFEIPGSEISSVHVHEGCVSRGEPPRTARRAAAPAPAPRDPHLHDWPELRITNCTYCYNPSVKSPNQAARSAFPSPPVPSAQPT
Summary
Uniprot
A0A2A4JII0
A0A2A4JH32
A0A2H1V364
A0A194QFX1
A0A212EVP6
A0A2W1BDB6
+ More
A0A0M9A6F5 A0A154PSU5 A0A0P5GLK4 A0A147B8V2 A0A2R5LGM9 A0A0P5I0E5 A0A151I4E0 A0A158N9I1 L7M0G7 L7M2K0 E0VRL3 A0A0P5S3L8 A0A131XGR3 A0A1B6D567 A0A087TU49 A0A0P5FU31 A0A0P5I5F1 A0A0P6EA02 A0A0P5S3M1 A0A0P5RY93 A0A0P5K1B9 A0A0P5RJX8 A0A0P5HUD0 A0A0P5JW54 A0A0P6D511 A0A0P5IWS9 A0A0N8CB83 A0A0P5HD23 A0A0P5GY51 A0A0N8C5B0 A0A0P5LVU1 A0A2C9JKK1 A0A0P5EQU7 A0A0P5KPY0 A0A0P5KNN8 A0A0P6EFY2 A0A0P5RW34 A0A0P5RY21 A0A0P5GXW5 A0A0P6EYJ7 A0A0P5ISM0 A0A0P5VR28 A0A0P5X981 A0A0P5L5X7 A0A0P5AKS7 A0A0P4ZND5 A0A1E1X3K1 A0A0P5C5X0 A0A0P5T9P2 A0A0P5SJ64 A0A224YI54 I3M2X9 A0A2C9JKK9 W5LEJ7 A0A131YJZ6 H2Q9L9 A0A2R9C0Z7 A0A3B4BMW0 A0A2K6BJT5 A0A2K5V0Y7 F7HHL5 A0A2Y9K401 A0A0P5JNA1 A0A0P7Z9Z4 A0A0P5JFG1 A0A0P6FW94 A0A1U7QMZ9 A0A2Y9NK87 A0A2K5NI46 F1SJL5 A0A383YTG9 A0A452FV43 V5I522 A0A2K6PN89 A0A2K5HY01 A0A0B7B8S8 A0A0P5RTR5 A0A0P6H5W5 A0A3L7HV47 A0A2K6PN40 A0A0P5NTI3 A0A2K6PN46 A0A2K5HY84 A0A452HAT2 A0A2K5HY54 H0V0S0 A0A2H6N419 A0A0P5K5I4 A0A2U3Z5M0 M7BV10 A0A1W5AZ15 G1RQ25 Q4V866 A0A1L8H1C6
A0A0M9A6F5 A0A154PSU5 A0A0P5GLK4 A0A147B8V2 A0A2R5LGM9 A0A0P5I0E5 A0A151I4E0 A0A158N9I1 L7M0G7 L7M2K0 E0VRL3 A0A0P5S3L8 A0A131XGR3 A0A1B6D567 A0A087TU49 A0A0P5FU31 A0A0P5I5F1 A0A0P6EA02 A0A0P5S3M1 A0A0P5RY93 A0A0P5K1B9 A0A0P5RJX8 A0A0P5HUD0 A0A0P5JW54 A0A0P6D511 A0A0P5IWS9 A0A0N8CB83 A0A0P5HD23 A0A0P5GY51 A0A0N8C5B0 A0A0P5LVU1 A0A2C9JKK1 A0A0P5EQU7 A0A0P5KPY0 A0A0P5KNN8 A0A0P6EFY2 A0A0P5RW34 A0A0P5RY21 A0A0P5GXW5 A0A0P6EYJ7 A0A0P5ISM0 A0A0P5VR28 A0A0P5X981 A0A0P5L5X7 A0A0P5AKS7 A0A0P4ZND5 A0A1E1X3K1 A0A0P5C5X0 A0A0P5T9P2 A0A0P5SJ64 A0A224YI54 I3M2X9 A0A2C9JKK9 W5LEJ7 A0A131YJZ6 H2Q9L9 A0A2R9C0Z7 A0A3B4BMW0 A0A2K6BJT5 A0A2K5V0Y7 F7HHL5 A0A2Y9K401 A0A0P5JNA1 A0A0P7Z9Z4 A0A0P5JFG1 A0A0P6FW94 A0A1U7QMZ9 A0A2Y9NK87 A0A2K5NI46 F1SJL5 A0A383YTG9 A0A452FV43 V5I522 A0A2K6PN89 A0A2K5HY01 A0A0B7B8S8 A0A0P5RTR5 A0A0P6H5W5 A0A3L7HV47 A0A2K6PN40 A0A0P5NTI3 A0A2K6PN46 A0A2K5HY84 A0A452HAT2 A0A2K5HY54 H0V0S0 A0A2H6N419 A0A0P5K5I4 A0A2U3Z5M0 M7BV10 A0A1W5AZ15 G1RQ25 Q4V866 A0A1L8H1C6
Pubmed
EMBL
NWSH01001396
PCG71384.1
PCG71385.1
ODYU01000454
SOQ35285.1
KQ458981
+ More
KPJ04448.1 AGBW02012143 OWR45568.1 KZ150141 PZC72948.1 KQ435737 KOX76893.1 KQ435066 KZC14328.1 GDIQ01240749 JAK10976.1 GEIB01001417 JAR86815.1 GGLE01004557 MBY08683.1 GDIQ01220200 JAK31525.1 KQ976461 KYM84712.1 ADTU01009655 GACK01007667 JAA57367.1 GACK01007666 JAA57368.1 DS235478 EEB16019.1 GDIQ01092837 JAL58889.1 GEFH01002278 JAP66303.1 GEDC01016456 JAS20842.1 KK116737 KFM68638.1 GDIQ01250289 JAK01436.1 GDIQ01223198 JAK28527.1 GDIQ01065655 JAN29082.1 GDIQ01092838 JAL58888.1 GDIQ01099696 JAL52030.1 GDIQ01216848 JAK34877.1 GDIQ01099697 JAL52029.1 GDIQ01223197 JAK28528.1 GDIQ01198161 GDIQ01166483 GDIQ01050312 JAK53564.1 GDIQ01082685 JAN12052.1 GDIQ01206656 JAK45069.1 GDIQ01096795 JAL54931.1 GDIQ01235449 JAK16276.1 GDIQ01235448 JAK16277.1 GDIQ01113379 JAL38347.1 GDIQ01188898 JAK62827.1 GDIQ01266116 JAJ85608.1 GDIQ01182319 JAK69406.1 GDIQ01188900 JAK62825.1 GDIQ01064251 JAN30486.1 GDIQ01113378 JAL38348.1 GDIQ01094985 JAL56741.1 GDIQ01234295 JAK17430.1 GDIQ01068502 JAN26235.1 GDIQ01234293 JAK17432.1 GDIP01096600 JAM07115.1 GDIP01087606 JAM16109.1 GDIQ01174504 JAK77221.1 GDIP01211333 JAJ12069.1 GDIP01210230 JAJ13172.1 GFAC01005338 JAT93850.1 GDIP01179332 JAJ44070.1 GDIQ01094983 JAL56743.1 GDIQ01094984 JAL56742.1 GFPF01006201 MAA17347.1 AGTP01003211 AGTP01003212 AGTP01003213 GEDV01009290 JAP79267.1 AACZ04064621 AJFE02098759 AJFE02098760 AJFE02098761 AJFE02098762 AJFE02098763 AQIA01063011 AQIA01063012 AQIA01063013 JSUE03038534 JSUE03038535 GDIQ01200761 JAK50964.1 JARO02000728 KPP77589.1 GDIQ01200760 JAK50965.1 GDIQ01043457 JAN51280.1 AEMK02000004 DQIR01223145 DQIR01262373 DQIR01310459 HDB78622.1 LWLT01000010 GANP01000735 JAB83733.1 HACG01041587 HACG01041590 CEK88452.1 CEK88455.1 GDIQ01096796 GDIQ01050313 JAL54930.1 GDIQ01045616 JAN49121.1 RAZU01000152 RLQ69221.1 GDIQ01142130 JAL09596.1 AAKN02051087 AAKN02051088 IACI01047124 LAA24099.1 GDIQ01188901 JAK62824.1 KB486406 EMP41671.1 ADFV01035763 ADFV01035764 ADFV01035765 ADFV01035766 ADFV01035767 BC097522 AAH97522.1 CM004470 OCT89811.1
KPJ04448.1 AGBW02012143 OWR45568.1 KZ150141 PZC72948.1 KQ435737 KOX76893.1 KQ435066 KZC14328.1 GDIQ01240749 JAK10976.1 GEIB01001417 JAR86815.1 GGLE01004557 MBY08683.1 GDIQ01220200 JAK31525.1 KQ976461 KYM84712.1 ADTU01009655 GACK01007667 JAA57367.1 GACK01007666 JAA57368.1 DS235478 EEB16019.1 GDIQ01092837 JAL58889.1 GEFH01002278 JAP66303.1 GEDC01016456 JAS20842.1 KK116737 KFM68638.1 GDIQ01250289 JAK01436.1 GDIQ01223198 JAK28527.1 GDIQ01065655 JAN29082.1 GDIQ01092838 JAL58888.1 GDIQ01099696 JAL52030.1 GDIQ01216848 JAK34877.1 GDIQ01099697 JAL52029.1 GDIQ01223197 JAK28528.1 GDIQ01198161 GDIQ01166483 GDIQ01050312 JAK53564.1 GDIQ01082685 JAN12052.1 GDIQ01206656 JAK45069.1 GDIQ01096795 JAL54931.1 GDIQ01235449 JAK16276.1 GDIQ01235448 JAK16277.1 GDIQ01113379 JAL38347.1 GDIQ01188898 JAK62827.1 GDIQ01266116 JAJ85608.1 GDIQ01182319 JAK69406.1 GDIQ01188900 JAK62825.1 GDIQ01064251 JAN30486.1 GDIQ01113378 JAL38348.1 GDIQ01094985 JAL56741.1 GDIQ01234295 JAK17430.1 GDIQ01068502 JAN26235.1 GDIQ01234293 JAK17432.1 GDIP01096600 JAM07115.1 GDIP01087606 JAM16109.1 GDIQ01174504 JAK77221.1 GDIP01211333 JAJ12069.1 GDIP01210230 JAJ13172.1 GFAC01005338 JAT93850.1 GDIP01179332 JAJ44070.1 GDIQ01094983 JAL56743.1 GDIQ01094984 JAL56742.1 GFPF01006201 MAA17347.1 AGTP01003211 AGTP01003212 AGTP01003213 GEDV01009290 JAP79267.1 AACZ04064621 AJFE02098759 AJFE02098760 AJFE02098761 AJFE02098762 AJFE02098763 AQIA01063011 AQIA01063012 AQIA01063013 JSUE03038534 JSUE03038535 GDIQ01200761 JAK50964.1 JARO02000728 KPP77589.1 GDIQ01200760 JAK50965.1 GDIQ01043457 JAN51280.1 AEMK02000004 DQIR01223145 DQIR01262373 DQIR01310459 HDB78622.1 LWLT01000010 GANP01000735 JAB83733.1 HACG01041587 HACG01041590 CEK88452.1 CEK88455.1 GDIQ01096796 GDIQ01050313 JAL54930.1 GDIQ01045616 JAN49121.1 RAZU01000152 RLQ69221.1 GDIQ01142130 JAL09596.1 AAKN02051087 AAKN02051088 IACI01047124 LAA24099.1 GDIQ01188901 JAK62824.1 KB486406 EMP41671.1 ADFV01035763 ADFV01035764 ADFV01035765 ADFV01035766 ADFV01035767 BC097522 AAH97522.1 CM004470 OCT89811.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053105
UP000076502
UP000078540
+ More
UP000005205 UP000009046 UP000054359 UP000076420 UP000005215 UP000018467 UP000002277 UP000240080 UP000261440 UP000233120 UP000233100 UP000006718 UP000248482 UP000034805 UP000189706 UP000248483 UP000233060 UP000008227 UP000261681 UP000291000 UP000233200 UP000233080 UP000273346 UP000291020 UP000005447 UP000245341 UP000031443 UP000192224 UP000001073 UP000186698
UP000005205 UP000009046 UP000054359 UP000076420 UP000005215 UP000018467 UP000002277 UP000240080 UP000261440 UP000233120 UP000233100 UP000006718 UP000248482 UP000034805 UP000189706 UP000248483 UP000233060 UP000008227 UP000261681 UP000291000 UP000233200 UP000233080 UP000273346 UP000291020 UP000005447 UP000245341 UP000031443 UP000192224 UP000001073 UP000186698
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2A4JII0
A0A2A4JH32
A0A2H1V364
A0A194QFX1
A0A212EVP6
A0A2W1BDB6
+ More
A0A0M9A6F5 A0A154PSU5 A0A0P5GLK4 A0A147B8V2 A0A2R5LGM9 A0A0P5I0E5 A0A151I4E0 A0A158N9I1 L7M0G7 L7M2K0 E0VRL3 A0A0P5S3L8 A0A131XGR3 A0A1B6D567 A0A087TU49 A0A0P5FU31 A0A0P5I5F1 A0A0P6EA02 A0A0P5S3M1 A0A0P5RY93 A0A0P5K1B9 A0A0P5RJX8 A0A0P5HUD0 A0A0P5JW54 A0A0P6D511 A0A0P5IWS9 A0A0N8CB83 A0A0P5HD23 A0A0P5GY51 A0A0N8C5B0 A0A0P5LVU1 A0A2C9JKK1 A0A0P5EQU7 A0A0P5KPY0 A0A0P5KNN8 A0A0P6EFY2 A0A0P5RW34 A0A0P5RY21 A0A0P5GXW5 A0A0P6EYJ7 A0A0P5ISM0 A0A0P5VR28 A0A0P5X981 A0A0P5L5X7 A0A0P5AKS7 A0A0P4ZND5 A0A1E1X3K1 A0A0P5C5X0 A0A0P5T9P2 A0A0P5SJ64 A0A224YI54 I3M2X9 A0A2C9JKK9 W5LEJ7 A0A131YJZ6 H2Q9L9 A0A2R9C0Z7 A0A3B4BMW0 A0A2K6BJT5 A0A2K5V0Y7 F7HHL5 A0A2Y9K401 A0A0P5JNA1 A0A0P7Z9Z4 A0A0P5JFG1 A0A0P6FW94 A0A1U7QMZ9 A0A2Y9NK87 A0A2K5NI46 F1SJL5 A0A383YTG9 A0A452FV43 V5I522 A0A2K6PN89 A0A2K5HY01 A0A0B7B8S8 A0A0P5RTR5 A0A0P6H5W5 A0A3L7HV47 A0A2K6PN40 A0A0P5NTI3 A0A2K6PN46 A0A2K5HY84 A0A452HAT2 A0A2K5HY54 H0V0S0 A0A2H6N419 A0A0P5K5I4 A0A2U3Z5M0 M7BV10 A0A1W5AZ15 G1RQ25 Q4V866 A0A1L8H1C6
A0A0M9A6F5 A0A154PSU5 A0A0P5GLK4 A0A147B8V2 A0A2R5LGM9 A0A0P5I0E5 A0A151I4E0 A0A158N9I1 L7M0G7 L7M2K0 E0VRL3 A0A0P5S3L8 A0A131XGR3 A0A1B6D567 A0A087TU49 A0A0P5FU31 A0A0P5I5F1 A0A0P6EA02 A0A0P5S3M1 A0A0P5RY93 A0A0P5K1B9 A0A0P5RJX8 A0A0P5HUD0 A0A0P5JW54 A0A0P6D511 A0A0P5IWS9 A0A0N8CB83 A0A0P5HD23 A0A0P5GY51 A0A0N8C5B0 A0A0P5LVU1 A0A2C9JKK1 A0A0P5EQU7 A0A0P5KPY0 A0A0P5KNN8 A0A0P6EFY2 A0A0P5RW34 A0A0P5RY21 A0A0P5GXW5 A0A0P6EYJ7 A0A0P5ISM0 A0A0P5VR28 A0A0P5X981 A0A0P5L5X7 A0A0P5AKS7 A0A0P4ZND5 A0A1E1X3K1 A0A0P5C5X0 A0A0P5T9P2 A0A0P5SJ64 A0A224YI54 I3M2X9 A0A2C9JKK9 W5LEJ7 A0A131YJZ6 H2Q9L9 A0A2R9C0Z7 A0A3B4BMW0 A0A2K6BJT5 A0A2K5V0Y7 F7HHL5 A0A2Y9K401 A0A0P5JNA1 A0A0P7Z9Z4 A0A0P5JFG1 A0A0P6FW94 A0A1U7QMZ9 A0A2Y9NK87 A0A2K5NI46 F1SJL5 A0A383YTG9 A0A452FV43 V5I522 A0A2K6PN89 A0A2K5HY01 A0A0B7B8S8 A0A0P5RTR5 A0A0P6H5W5 A0A3L7HV47 A0A2K6PN40 A0A0P5NTI3 A0A2K6PN46 A0A2K5HY84 A0A452HAT2 A0A2K5HY54 H0V0S0 A0A2H6N419 A0A0P5K5I4 A0A2U3Z5M0 M7BV10 A0A1W5AZ15 G1RQ25 Q4V866 A0A1L8H1C6
PDB
1UM8
E-value=5.11941e-80,
Score=759
Ontologies
GO
PANTHER
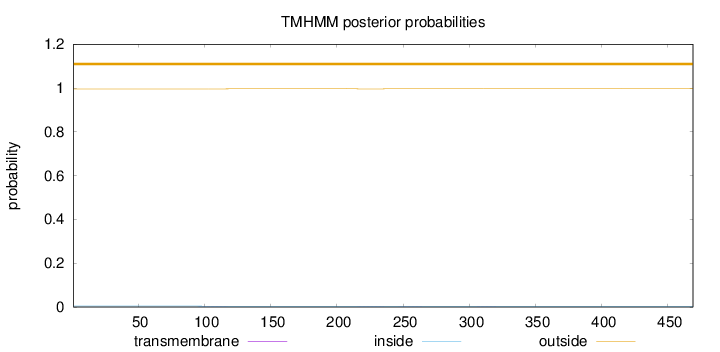
Topology
Length:
469
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02566
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00429
outside
1 - 469
Population Genetic Test Statistics
Pi
28.843132
Theta
24.932934
Tajima's D
-1.548942
CLR
0.754033
CSRT
0.0525973701314934
Interpretation
Uncertain
