Pre Gene Modal
BGIBMGA007210
Annotation
PREDICTED:_ATP-dependent_Clp_protease_ATP-binding_subunit_clpX-like?_mitochondrial_[Papilio_polytes]
Location in the cell
Extracellular Reliability : 1.898
Sequence
CDS
ATGAGTAGTGTAAGATTAAGTTTTGTGTCCGTGGGAAGGCTAGCGGTGAGAAGAAACCCACAGATAACATCAGGTGTCCAGCAGTGTCGCTCCCTCAGACGCACGTGCACCAGAGCGCTCGGTACCTGCACAGTACTATGCAAGAGTCCAACAGAACAGCCTCCGGCGCCTCCGCCATCTAACTCAGGGAAAGATGCCGCTGCCTCAGGCACATCGGGTGGAAAAAAGTCTGGCAACGGTGGTAACGGAGTACTGACATGTCCGAAATGCGGAGACCCCTGTACGCACGTCGAGACCTTCGTGAGTTCGACGCGTTTCGTGAAGTGCGACAAGTGCCACCATTTCTTCGTGGTTCTCAGTGAAGTGGACACCAAGAAGAGTATTAAAGATAGTACAGACAACAAATCAGGGTTCTACAGGTAA
Protein
MSSVRLSFVSVGRLAVRRNPQITSGVQQCRSLRRTCTRALGTCTVLCKSPTEQPPAPPPSNSGKDAAASGTSGGKKSGNGGNGVLTCPKCGDPCTHVETFVSSTRFVKCDKCHHFFVVLSEVDTKKSIKDSTDNKSGFYR
Summary
Uniprot
Pubmed
EMBL
NWSH01001396
PCG71384.1
ODYU01000454
SOQ35285.1
AGBW02012143
OWR45568.1
+ More
KQ459995 KPJ18512.1 RSAL01000117 RVE46817.1 KQ458981 KPJ04448.1 KQ435066 KZC14328.1 KQ971311 KYB29493.1 KK852756 KDR17084.1 KK107260 EZA54198.1 KQ414868 KOC59975.1 LBMM01000816 KMQ97421.1 GL766449 EFZ15206.1 JR040561 JR040562 AEY59183.1 AEY59184.1 GL888628 EGI58750.1 NNAY01000309 OXU29311.1 KQ977545 KYN02034.1 BT128013 AEE62975.1 APGK01016968 KB739995 KB630343 KB631518 ENN82007.1 ERL83542.1 ERL84345.1 GL449444 EFN82674.1 GALX01004219 JAB64247.1 GEZM01083903 JAV60961.1 GEZM01083905 JAV60958.1 KQ982169 KYQ59461.1
KQ459995 KPJ18512.1 RSAL01000117 RVE46817.1 KQ458981 KPJ04448.1 KQ435066 KZC14328.1 KQ971311 KYB29493.1 KK852756 KDR17084.1 KK107260 EZA54198.1 KQ414868 KOC59975.1 LBMM01000816 KMQ97421.1 GL766449 EFZ15206.1 JR040561 JR040562 AEY59183.1 AEY59184.1 GL888628 EGI58750.1 NNAY01000309 OXU29311.1 KQ977545 KYN02034.1 BT128013 AEE62975.1 APGK01016968 KB739995 KB630343 KB631518 ENN82007.1 ERL83542.1 ERL84345.1 GL449444 EFN82674.1 GALX01004219 JAB64247.1 GEZM01083903 JAV60961.1 GEZM01083905 JAV60958.1 KQ982169 KYQ59461.1
Proteomes
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
Ontologies
PANTHER
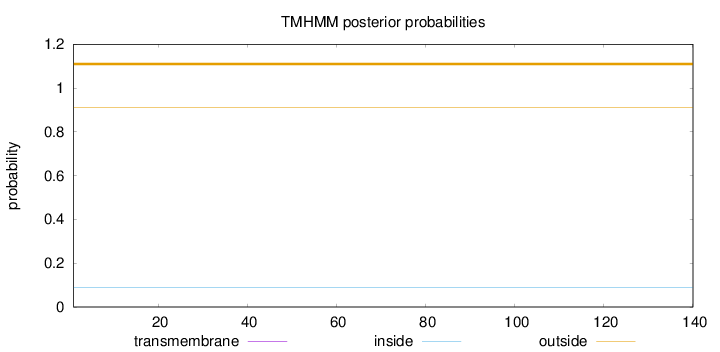
Topology
Length:
140
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08866
outside
1 - 140
Population Genetic Test Statistics
Pi
21.191124
Theta
23.187413
Tajima's D
-0.676406
CLR
10.852803
CSRT
0.202689865506725
Interpretation
Uncertain
