Pre Gene Modal
BGIBMGA007147
Annotation
Sorting_nexin-27_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 1.641
Sequence
CDS
ATGGCTGAGACGGAAACCGATAATAGTAGTGTAGTTAGAGCGGAGAGAAACAACAAGAGCAATAACAATGGCAACGGGCCACGACGCGTTACTATATACAAAACGGAGACCGGGTTTGGTTTTAACGTCCGCGGCCAAGTGTCTGAGGGTGGACAGCTAAGATCGATCAACGGGGAATTGTATGCTCCTCTGCAGCATGTCAGCGCCGTGCTGGAGAACGGAGCGGCTGAACAGGCCGGCATCAGAAAAGGAGACCGAATATTAGAAGTGTGCGTATTCAAACCTCTCCATCAATTTATTGTTTTTAATTTAATATGTTTCAAGAGGTCAAATTTAAATGAAATAAAATTCTTAAAAATAATTCATGCTATCTTCTCTATTCCGATTAACTAA
Protein
MAETETDNSSVVRAERNNKSNNNGNGPRRVTIYKTETGFGFNVRGQVSEGGQLRSINGELYAPLQHVSAVLENGAAEQAGIRKGDRILEVCVFKPLHQFIVFNLICFKRSNLNEIKFLKIIHAIFSIPIN
Summary
Uniprot
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
5EMB
E-value=1.81039e-24,
Score=272
Ontologies
GO
PANTHER
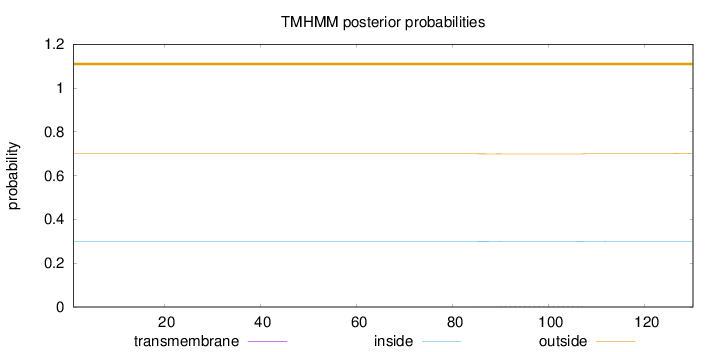
Topology
Length:
130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07788
Exp number, first 60 AAs:
0
Total prob of N-in:
0.30005
outside
1 - 130
Population Genetic Test Statistics
Pi
2.524358
Theta
13.262796
Tajima's D
-2.28246
CLR
142.705523
CSRT
0.00344982750862457
Interpretation
Possibly Positive selection
