Gene
KWMTBOMO12783
Pre Gene Modal
BGIBMGA007151
Annotation
PREDICTED:_uncharacterized_family_31_glucosidase_KIAA1161_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.416 Nuclear Reliability : 1.955
Sequence
CDS
ATGCCCAATTTTCTGTGCCTGTGTGAAGATAGAACCACATCACCCCTCAAATCCCCGCGCCGGAAGTCAGGCTACCCACGCCGGCTGGGCAGATCAGAATTCAGCATGGATGAAGATGATTACTCGCCGAGTAATTCTGTGACTTCTGTCAACAGCCTCGCCAGCCTCCTTAGGGAGAAGTTACAGAGTATTCCTCAGAAGATTCGTAAGAAACCATCAGACTACAAGCTCAAGGCGTTCGTGGCTCTAATGTTCCTGGCCGTGGCGTTTTTCATCGGCTTCGCGTATGTGCTCTATCACCAGCAGGCTCACTCCAAGACTTACTTCCGAAACGTTCAGCTGAACGAACCAAAGAGGCTAATAAGGATCTACAATGAGGACGATGTGGAAATACTGAAAGCTAACTTAGCGGTTGGCGTCCAAGGTAAAATAAAAGCCTTCCCATGTCTACCGGAAGACCGCCGCAACGGTTCCCAGTGTCTGGAGTGGCTCCACACCCTCCGTTTCTACATGAAGTCGCAGCCGCACGACCCTGGTTTGGACAACACAACATGCTACTCCATCCACTGGCAAGCGTTAAGACCTGATGTAACTCCTACGGACTGCTTCGACTGGGGTGACACAAAGGTTCACTGGTATGGCGGCGGACAGTCTTTTAACTTGACTTGGCCTCTGGACGCAGGCTCCATAAGCTTTACACCATTTGTTACCGGTGATATGCAGAAATCGCAATGGGGCAACCTCCTGACCAGGTACTTCATCAACACTAAAGGAGCAGCAATCATTGTCGACGAAGAAACTCCTTTACACGTAGCCGTCAAGCCTGACACCAAAGAAATATGCATTAGAGCTAAATACGACGACTTCGCTTTTGCCAACAAGATTACTGAATTTCCCGAAATGAGATACAACATTTGCACCTCAATCGATATGAAGTCGCTTCATACTTCCATTCACAGTCACAGGAGTTCACCTCTATGGGACGGTCTCAAGCCTGCCGACATGCAGACCTTAGATTCATTGATATCGGAACCGGTCTGGCAAATAGCGCCACGATTCAAACACGAACTTCAAGCGGAAACCATTGCTAAGTACACCGAGGACGTCATAAACTTAGGATTCTTAAAACAAGGGCATGTGCTCATTAACGAACATTGGCAAAATGAAATCGGTGATCTAGAAGTTGACAAGAGCCGTTTTGAAACTTTAAATGTGACTGTGGACAAATTGCATCGAAGAGGTTTTAAGGTGGCGTTCACTATCCAACCATTCATAAGCACGGAGAGCCAGAATTTCGCTGAGACAGTCCAGAAGCGGTTGCTAGGGTTTTAG
Protein
MPNFLCLCEDRTTSPLKSPRRKSGYPRRLGRSEFSMDEDDYSPSNSVTSVNSLASLLREKLQSIPQKIRKKPSDYKLKAFVALMFLAVAFFIGFAYVLYHQQAHSKTYFRNVQLNEPKRLIRIYNEDDVEILKANLAVGVQGKIKAFPCLPEDRRNGSQCLEWLHTLRFYMKSQPHDPGLDNTTCYSIHWQALRPDVTPTDCFDWGDTKVHWYGGGQSFNLTWPLDAGSISFTPFVTGDMQKSQWGNLLTRYFINTKGAAIIVDEETPLHVAVKPDTKEICIRAKYDDFAFANKITEFPEMRYNICTSIDMKSLHTSIHSHRSSPLWDGLKPADMQTLDSLISEPVWQIAPRFKHELQAETIAKYTEDVINLGFLKQGHVLINEHWQNEIGDLEVDKSRFETLNVTVDKLHRRGFKVAFTIQPFISTESQNFAETVQKRLLGF
Summary
Similarity
Belongs to the glycosyl hydrolase 31 family.
Uniprot
H9JCA6
A0A2A4JPW7
A0A2W1BF92
A0A2H1V9Q2
A0A437B8Y9
A0A194QG35
+ More
A0A194RKZ5 A0A2A4JNX2 A0A212EVP2 A0A0L7LTR3 D6W9J2 A0A139WMW5 N6U046 V5G368 A0A1W4W7I6 A0A1W4WIW6 A0A1W4WJ51 A0A1Y1LC80 A0A1Y1LC30 A0A1Y1LEA5 K7IMG7 E0VB12 A0A232EYT2 A0A126CSW8 A0A146KWN4 A0A0K8SZG3 A0A0K8T0D7 A0A146MAG2 A0A158P0U2 E9J762 A0A1B6KUE2 A0A067R3Y5 A0A0A9XW03 A0A1B6E3L4 A0A1B6ESZ6 A0A1B6CVS0 A0A151I962 A0A026WB22 A0A0L0C1Q4 A0A2J7QIU2 A0A0C9R4F4 A0A158P0T4 A0A195B5W0 A0A0C9RDI2 F4W984 A0A158P0S6 A0A2J7QIV9 T1HFB1 A0A3L8DQH0 A0A1A9W7A4 E2AQK9 A0A0T6B3V5 A0A224X761 A0A154P8S5 A0A1A9UZN7 E2BP96 A0A1A9YI79 A0A2J7QIT9 A0A1B0BK38 A0A195EEM5 A0A195F3Y7 A0A151WKM3 A0A1I8MW13 A0A0L7RI03 T1PDI3 A0A0K8TSZ7 W8BR02 A0A088AQ74 A0A1B0AHW3 Q179V8 F5HMZ1 A0A182XNW9 A0A182V1B0 W8C1T3 F5HMZ0 W8B3G2 A0A182SLU8 A0A1B0GEH1 A0A182JXR2 A0A182VWA2 A0A023EW00 A0A182L891 A0A182HK16 A0A182UKD3 Q7PWY6 A0A182MJV1 A0A1Q3FZ57 A0A034VBK5 J9K3K3 A0A1I8MWD4 A0A1I8PA06 A0A2P8YIN3 A0A182R543 A0A0A1WRN8 A0A310SJ49 A0A182IKR9 A0A034VF04 A0A2H8TR74 A0A0N0BI36 A0A1L8DQV8
A0A194RKZ5 A0A2A4JNX2 A0A212EVP2 A0A0L7LTR3 D6W9J2 A0A139WMW5 N6U046 V5G368 A0A1W4W7I6 A0A1W4WIW6 A0A1W4WJ51 A0A1Y1LC80 A0A1Y1LC30 A0A1Y1LEA5 K7IMG7 E0VB12 A0A232EYT2 A0A126CSW8 A0A146KWN4 A0A0K8SZG3 A0A0K8T0D7 A0A146MAG2 A0A158P0U2 E9J762 A0A1B6KUE2 A0A067R3Y5 A0A0A9XW03 A0A1B6E3L4 A0A1B6ESZ6 A0A1B6CVS0 A0A151I962 A0A026WB22 A0A0L0C1Q4 A0A2J7QIU2 A0A0C9R4F4 A0A158P0T4 A0A195B5W0 A0A0C9RDI2 F4W984 A0A158P0S6 A0A2J7QIV9 T1HFB1 A0A3L8DQH0 A0A1A9W7A4 E2AQK9 A0A0T6B3V5 A0A224X761 A0A154P8S5 A0A1A9UZN7 E2BP96 A0A1A9YI79 A0A2J7QIT9 A0A1B0BK38 A0A195EEM5 A0A195F3Y7 A0A151WKM3 A0A1I8MW13 A0A0L7RI03 T1PDI3 A0A0K8TSZ7 W8BR02 A0A088AQ74 A0A1B0AHW3 Q179V8 F5HMZ1 A0A182XNW9 A0A182V1B0 W8C1T3 F5HMZ0 W8B3G2 A0A182SLU8 A0A1B0GEH1 A0A182JXR2 A0A182VWA2 A0A023EW00 A0A182L891 A0A182HK16 A0A182UKD3 Q7PWY6 A0A182MJV1 A0A1Q3FZ57 A0A034VBK5 J9K3K3 A0A1I8MWD4 A0A1I8PA06 A0A2P8YIN3 A0A182R543 A0A0A1WRN8 A0A310SJ49 A0A182IKR9 A0A034VF04 A0A2H8TR74 A0A0N0BI36 A0A1L8DQV8
Pubmed
19121390
28756777
26354079
22118469
26227816
18362917
+ More
19820115 23537049 28004739 20075255 20566863 28648823 26823975 21347285 21282665 24845553 25401762 24508170 26108605 21719571 30249741 20798317 25315136 26369729 24495485 17510324 12364791 14747013 17210077 24945155 20966253 25348373 29403074 25830018
19820115 23537049 28004739 20075255 20566863 28648823 26823975 21347285 21282665 24845553 25401762 24508170 26108605 21719571 30249741 20798317 25315136 26369729 24495485 17510324 12364791 14747013 17210077 24945155 20966253 25348373 29403074 25830018
EMBL
BABH01031633
BABH01031634
NWSH01000878
PCG73748.1
KZ150141
PZC72951.1
+ More
ODYU01001417 SOQ37573.1 RSAL01000117 RVE46819.1 KQ458981 KPJ04452.1 KQ459995 KPJ18508.1 PCG73747.1 AGBW02012143 OWR45565.1 JTDY01000099 KOB78873.1 KQ971312 EEZ98146.2 KYB29206.1 APGK01044428 KB741026 KB631924 ENN74915.1 ERL87164.1 GALX01004021 JAB64445.1 GEZM01060171 JAV71223.1 GEZM01060172 JAV71222.1 GEZM01060173 JAV71221.1 DS235020 EEB10568.1 NNAY01001605 OXU23447.1 KC702766 AHG54238.1 GDHC01019053 JAP99575.1 GBRD01007182 JAG58639.1 GBRD01007181 JAG58640.1 GDHC01004694 GDHC01002863 JAQ13935.1 JAQ15766.1 ADTU01001023 ADTU01001024 ADTU01001025 GL768421 EFZ11396.1 GEBQ01024924 JAT15053.1 KK852913 KDR13896.1 GBHO01020556 JAG23048.1 GEDC01004776 JAS32522.1 GECZ01028644 JAS41125.1 GEDC01019780 JAS17518.1 KQ978323 KYM95139.1 KK107293 EZA53260.1 JRES01001001 KNC26235.1 NEVH01013577 PNF28508.1 GBYB01011179 JAG80946.1 KQ976598 KYM79579.1 GBYB01011177 JAG80944.1 GL888002 EGI69266.1 PNF28506.1 ACPB03021649 QOIP01000005 RLU22526.1 GL441777 EFN64277.1 LJIG01009940 KRT82076.1 GFTR01008129 JAW08297.1 KQ434829 KZC07764.1 GL449553 EFN82516.1 PNF28507.1 JXJN01015782 KQ979039 KYN23257.1 KQ981855 KYN34887.1 KQ983002 KYQ48408.1 KQ414590 KOC70356.1 KA646754 AFP61383.1 GDAI01000121 JAI17482.1 GAMC01010899 JAB95656.1 CH477344 EAT43040.1 AAAB01008987 EGK97662.1 GAMC01010901 JAB95654.1 EGK97663.1 GAMC01010902 GAMC01010900 JAB95655.1 CCAG010012258 CCAG010012259 GAPW01000327 JAC13271.1 APCN01000814 EAA00985.4 AXCM01001932 GFDL01002201 JAV32844.1 GAKP01018281 GAKP01018277 JAC40671.1 ABLF02026801 PYGN01000570 PSN44064.1 GBXI01013189 GBXI01012679 JAD01103.1 JAD01613.1 KQ766694 OAD53590.1 GAKP01018285 GAKP01018271 JAC40667.1 GFXV01004297 MBW16102.1 KQ435739 KOX76864.1 GFDF01005320 JAV08764.1
ODYU01001417 SOQ37573.1 RSAL01000117 RVE46819.1 KQ458981 KPJ04452.1 KQ459995 KPJ18508.1 PCG73747.1 AGBW02012143 OWR45565.1 JTDY01000099 KOB78873.1 KQ971312 EEZ98146.2 KYB29206.1 APGK01044428 KB741026 KB631924 ENN74915.1 ERL87164.1 GALX01004021 JAB64445.1 GEZM01060171 JAV71223.1 GEZM01060172 JAV71222.1 GEZM01060173 JAV71221.1 DS235020 EEB10568.1 NNAY01001605 OXU23447.1 KC702766 AHG54238.1 GDHC01019053 JAP99575.1 GBRD01007182 JAG58639.1 GBRD01007181 JAG58640.1 GDHC01004694 GDHC01002863 JAQ13935.1 JAQ15766.1 ADTU01001023 ADTU01001024 ADTU01001025 GL768421 EFZ11396.1 GEBQ01024924 JAT15053.1 KK852913 KDR13896.1 GBHO01020556 JAG23048.1 GEDC01004776 JAS32522.1 GECZ01028644 JAS41125.1 GEDC01019780 JAS17518.1 KQ978323 KYM95139.1 KK107293 EZA53260.1 JRES01001001 KNC26235.1 NEVH01013577 PNF28508.1 GBYB01011179 JAG80946.1 KQ976598 KYM79579.1 GBYB01011177 JAG80944.1 GL888002 EGI69266.1 PNF28506.1 ACPB03021649 QOIP01000005 RLU22526.1 GL441777 EFN64277.1 LJIG01009940 KRT82076.1 GFTR01008129 JAW08297.1 KQ434829 KZC07764.1 GL449553 EFN82516.1 PNF28507.1 JXJN01015782 KQ979039 KYN23257.1 KQ981855 KYN34887.1 KQ983002 KYQ48408.1 KQ414590 KOC70356.1 KA646754 AFP61383.1 GDAI01000121 JAI17482.1 GAMC01010899 JAB95656.1 CH477344 EAT43040.1 AAAB01008987 EGK97662.1 GAMC01010901 JAB95654.1 EGK97663.1 GAMC01010902 GAMC01010900 JAB95655.1 CCAG010012258 CCAG010012259 GAPW01000327 JAC13271.1 APCN01000814 EAA00985.4 AXCM01001932 GFDL01002201 JAV32844.1 GAKP01018281 GAKP01018277 JAC40671.1 ABLF02026801 PYGN01000570 PSN44064.1 GBXI01013189 GBXI01012679 JAD01103.1 JAD01613.1 KQ766694 OAD53590.1 GAKP01018285 GAKP01018271 JAC40667.1 GFXV01004297 MBW16102.1 KQ435739 KOX76864.1 GFDF01005320 JAV08764.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000007266 UP000019118 UP000030742 UP000192223 UP000002358 UP000009046 UP000215335 UP000005205 UP000027135 UP000078542 UP000053097 UP000037069 UP000235965 UP000078540 UP000007755 UP000015103 UP000279307 UP000091820 UP000000311 UP000076502 UP000078200 UP000008237 UP000092443 UP000092460 UP000078492 UP000078541 UP000075809 UP000095301 UP000053825 UP000005203 UP000092445 UP000008820 UP000007062 UP000076407 UP000075903 UP000075901 UP000092444 UP000075881 UP000075920 UP000075882 UP000075840 UP000075902 UP000075883 UP000007819 UP000095300 UP000245037 UP000075900 UP000075880 UP000053105
UP000037510 UP000007266 UP000019118 UP000030742 UP000192223 UP000002358 UP000009046 UP000215335 UP000005205 UP000027135 UP000078542 UP000053097 UP000037069 UP000235965 UP000078540 UP000007755 UP000015103 UP000279307 UP000091820 UP000000311 UP000076502 UP000078200 UP000008237 UP000092443 UP000092460 UP000078492 UP000078541 UP000075809 UP000095301 UP000053825 UP000005203 UP000092445 UP000008820 UP000007062 UP000076407 UP000075903 UP000075901 UP000092444 UP000075881 UP000075920 UP000075882 UP000075840 UP000075902 UP000075883 UP000007819 UP000095300 UP000245037 UP000075900 UP000075880 UP000053105
PRIDE
Interpro
IPR000322
Glyco_hydro_31
+ More
IPR013780 Glyco_hydro_b
IPR017853 Glycoside_hydrolase_SF
IPR009057 Homeobox-like_sf
IPR036388 WH-like_DNA-bd_sf
IPR037833 SNX27_PX
IPR036871 PX_dom_sf
IPR037827 SNX27_FERM-like_dom
IPR001683 Phox
IPR000159 RA_dom
IPR041489 PDZ_6
IPR001478 PDZ
IPR036034 PDZ_sf
IPR018148 Methylglyoxal_synth_AS
IPR013780 Glyco_hydro_b
IPR017853 Glycoside_hydrolase_SF
IPR009057 Homeobox-like_sf
IPR036388 WH-like_DNA-bd_sf
IPR037833 SNX27_PX
IPR036871 PX_dom_sf
IPR037827 SNX27_FERM-like_dom
IPR001683 Phox
IPR000159 RA_dom
IPR041489 PDZ_6
IPR001478 PDZ
IPR036034 PDZ_sf
IPR018148 Methylglyoxal_synth_AS
Gene 3D
ProteinModelPortal
H9JCA6
A0A2A4JPW7
A0A2W1BF92
A0A2H1V9Q2
A0A437B8Y9
A0A194QG35
+ More
A0A194RKZ5 A0A2A4JNX2 A0A212EVP2 A0A0L7LTR3 D6W9J2 A0A139WMW5 N6U046 V5G368 A0A1W4W7I6 A0A1W4WIW6 A0A1W4WJ51 A0A1Y1LC80 A0A1Y1LC30 A0A1Y1LEA5 K7IMG7 E0VB12 A0A232EYT2 A0A126CSW8 A0A146KWN4 A0A0K8SZG3 A0A0K8T0D7 A0A146MAG2 A0A158P0U2 E9J762 A0A1B6KUE2 A0A067R3Y5 A0A0A9XW03 A0A1B6E3L4 A0A1B6ESZ6 A0A1B6CVS0 A0A151I962 A0A026WB22 A0A0L0C1Q4 A0A2J7QIU2 A0A0C9R4F4 A0A158P0T4 A0A195B5W0 A0A0C9RDI2 F4W984 A0A158P0S6 A0A2J7QIV9 T1HFB1 A0A3L8DQH0 A0A1A9W7A4 E2AQK9 A0A0T6B3V5 A0A224X761 A0A154P8S5 A0A1A9UZN7 E2BP96 A0A1A9YI79 A0A2J7QIT9 A0A1B0BK38 A0A195EEM5 A0A195F3Y7 A0A151WKM3 A0A1I8MW13 A0A0L7RI03 T1PDI3 A0A0K8TSZ7 W8BR02 A0A088AQ74 A0A1B0AHW3 Q179V8 F5HMZ1 A0A182XNW9 A0A182V1B0 W8C1T3 F5HMZ0 W8B3G2 A0A182SLU8 A0A1B0GEH1 A0A182JXR2 A0A182VWA2 A0A023EW00 A0A182L891 A0A182HK16 A0A182UKD3 Q7PWY6 A0A182MJV1 A0A1Q3FZ57 A0A034VBK5 J9K3K3 A0A1I8MWD4 A0A1I8PA06 A0A2P8YIN3 A0A182R543 A0A0A1WRN8 A0A310SJ49 A0A182IKR9 A0A034VF04 A0A2H8TR74 A0A0N0BI36 A0A1L8DQV8
A0A194RKZ5 A0A2A4JNX2 A0A212EVP2 A0A0L7LTR3 D6W9J2 A0A139WMW5 N6U046 V5G368 A0A1W4W7I6 A0A1W4WIW6 A0A1W4WJ51 A0A1Y1LC80 A0A1Y1LC30 A0A1Y1LEA5 K7IMG7 E0VB12 A0A232EYT2 A0A126CSW8 A0A146KWN4 A0A0K8SZG3 A0A0K8T0D7 A0A146MAG2 A0A158P0U2 E9J762 A0A1B6KUE2 A0A067R3Y5 A0A0A9XW03 A0A1B6E3L4 A0A1B6ESZ6 A0A1B6CVS0 A0A151I962 A0A026WB22 A0A0L0C1Q4 A0A2J7QIU2 A0A0C9R4F4 A0A158P0T4 A0A195B5W0 A0A0C9RDI2 F4W984 A0A158P0S6 A0A2J7QIV9 T1HFB1 A0A3L8DQH0 A0A1A9W7A4 E2AQK9 A0A0T6B3V5 A0A224X761 A0A154P8S5 A0A1A9UZN7 E2BP96 A0A1A9YI79 A0A2J7QIT9 A0A1B0BK38 A0A195EEM5 A0A195F3Y7 A0A151WKM3 A0A1I8MW13 A0A0L7RI03 T1PDI3 A0A0K8TSZ7 W8BR02 A0A088AQ74 A0A1B0AHW3 Q179V8 F5HMZ1 A0A182XNW9 A0A182V1B0 W8C1T3 F5HMZ0 W8B3G2 A0A182SLU8 A0A1B0GEH1 A0A182JXR2 A0A182VWA2 A0A023EW00 A0A182L891 A0A182HK16 A0A182UKD3 Q7PWY6 A0A182MJV1 A0A1Q3FZ57 A0A034VBK5 J9K3K3 A0A1I8MWD4 A0A1I8PA06 A0A2P8YIN3 A0A182R543 A0A0A1WRN8 A0A310SJ49 A0A182IKR9 A0A034VF04 A0A2H8TR74 A0A0N0BI36 A0A1L8DQV8
Ontologies
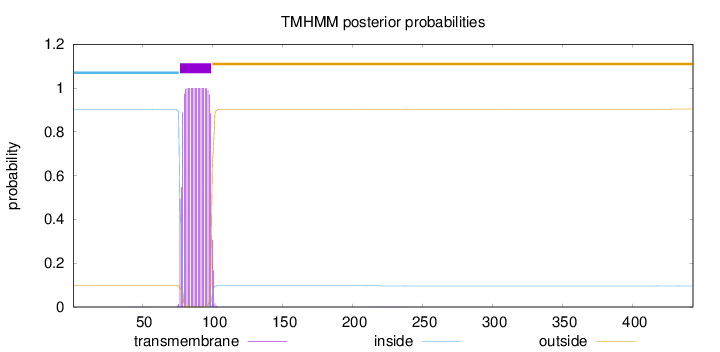
Topology
Length:
443
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.27864
Exp number, first 60 AAs:
0.00614
Total prob of N-in:
0.90278
inside
1 - 76
TMhelix
77 - 99
outside
100 - 443
Population Genetic Test Statistics
Pi
27.566485
Theta
24.003677
Tajima's D
0.340203
CLR
0.183969
CSRT
0.473126343682816
Interpretation
Uncertain
