Gene
KWMTBOMO12779
Annotation
hypothetical_protein_KGM_02824_[Danaus_plexippus]
Full name
Microtubule-associated protein futsch
+ More
Alpha-amylase
Alpha-amylase
Alternative Name
MAP1B homolog
Location in the cell
Cytoplasmic Reliability : 2.522
Sequence
CDS
ATGCTGCGGGGGCTGCTATCATGGGACGCTGGAGAGCACCAGGTCGATTTGGAGAAGGAACTTGCTACCCTTACGGCACAGGCACCGGAAGGGGAGGAAGCTCGCTATGGAGAGAGGCTGATCCAGTTCGCCTCGGAGAACCTGGTGACGGAGATCCTGATCCACCCGCAGATGAACACGCTGATGCAGTGCATGCGGAACCTGCTCAGCTCGTTCACGAGGCACAGGCACCTCGTGCACGCTGGATACACCTTCGCCGGAAACGGCTCCTGGATCATGCAAGACGGGACCTTCTCCCTGGCAGACTTCACAGACGCATACCAAGAGAACGAGGTCCAACGCGTTATTCGCGCATACGAGAACTCCATCTCGATCGATGTCCACTGCTCTACCAGCGGCGGGGGCGAGTGGGGCAGGCTCCCCGAGATGCCCTTCGTCAAATACTGCAAAATCAGGGTCAACCCTACTGATAAACTGGACCCTAGCTCTCAGGCCATCAAGGATTTTATCTTTAAACGTGACCTATTGGGCCCGAGGCATCCCCCATTACTTCCGTACCCTTTTATGTCTGTCTGA
Protein
MLRGLLSWDAGEHQVDLEKELATLTAQAPEGEEARYGERLIQFASENLVTEILIHPQMNTLMQCMRNLLSSFTRHRHLVHAGYTFAGNGSWIMQDGTFSLADFTDAYQENEVQRVIRAYENSISIDVHCSTSGGGEWGRLPEMPFVKYCKIRVNPTDKLDPSSQAIKDFIFKRDLLGPRHPPLLPYPFMSV
Summary
Description
During embryogenesis, necessary for dendritic and axonal organization and growth at the neuromuscular junction through the regulation of the synaptic microtubule cytoskeleton. Microtubule hairpin loops are found within a small subset of synaptic boutons at the neuromuscular synapse, these loops are stabilized by Futsch. Loop morphology and dynamics suggest that rearrangement of these microtubule-based loops is a critical component of the process of bouton division and for subsequent nerve-terminal growth and branching. Translation is repressed by Fmr1.
Subunit
Heterodimer of a heavy and a light chain. Interacts with Fmr1.
Miscellaneous
'Futsch' means 'gone' in German.
Similarity
Belongs to the glycosyl hydrolase 13 family.
Keywords
Complete proteome
Cytoplasm
Cytoskeleton
Developmental protein
Microtubule
Phosphoprotein
Reference proteome
Repeat
Feature
chain Microtubule-associated protein futsch
Uniprot
A0A212EMI4
A0A3S2TIC9
A0A2W1BJ16
A0A194QHP0
A0A194RR39
A0A0L7LU20
+ More
A0A2H1V5A6 A4IJ78 A0A1Y1KDM4 D6W9J1 A0A1I8N0Y2 W8BXP6 A0A1A9WCP8 A0A0A9WLA3 B3P9H8 A0A1B6IFC7 A0A1B0G446 A0A0R3NYE5 A0A0R3NYW6 Q29J01 B4PXP4 A0A1W4VS68 A0A1A9XAH7 M9NGG5 Q9W596 A0A0M3QYS9 A0A1A9ZRR8 A0A3B0KDE7 A0A1B0ATD1 A0A1A9UPH8 B4L480 A0A2P8YIH9 B4N1K5 A0A1I8PFQ4 B4MD01 A0A1B6DHQ9 A0A0L7RHR1 A0A088ACQ6 A0A1J1IZA5 A0A151I8Z5 A0A023EWM3 A0A195EDS2 A0A3L8DQ17 A0A195F2T2 A0A151WKN8 A0A158NDG7 A0A232EYR2 T1HAN9 A0A0J7L3P1 A0A0N0BDI5 K7IMG8 A0A154PLK1 A0A2Y9D0U7 A0A1Y9J0K7 A0A1S4GBB2 A0A182YHT9 A0A1B6M4Q5 Q0H6L5 Q0H6L4 Q0H6L3 Q0H6L6 A0A3R7SM39 B4I9E0 A0A336LLG9 B4GTL7 A0A0L0CN79 A0A0K2U468 B4JNT3 A0A2R7VZ47 U4TXE0 N6T9R7 E9J760 A0A0P5FZQ6 A0A182JSF9 A0A0P5JSG3 A0A2P2HW79 A0A0P5ENG7 A0A0P5FXU3 A0A0P6DMC4 A0A0P6G415 A0A0N7ZZC8 A0A0N8ATV5 A0A0P6E8R3 A0A0P6FFV1 A0A0N8DZ12 A0A0P5FWN8 A0A0P5JV90 A0A0P5WVF2 A0A0P4Y595 A0A0P5L9E6 A0A0P5W5G9 A0A0P5USX1 A0A0N8D0C2 A0A0P5JW72 A0A0P6GAG5 A0A0P6GCT4 A0A0P6EHX6 A0A0P5H8R7
A0A2H1V5A6 A4IJ78 A0A1Y1KDM4 D6W9J1 A0A1I8N0Y2 W8BXP6 A0A1A9WCP8 A0A0A9WLA3 B3P9H8 A0A1B6IFC7 A0A1B0G446 A0A0R3NYE5 A0A0R3NYW6 Q29J01 B4PXP4 A0A1W4VS68 A0A1A9XAH7 M9NGG5 Q9W596 A0A0M3QYS9 A0A1A9ZRR8 A0A3B0KDE7 A0A1B0ATD1 A0A1A9UPH8 B4L480 A0A2P8YIH9 B4N1K5 A0A1I8PFQ4 B4MD01 A0A1B6DHQ9 A0A0L7RHR1 A0A088ACQ6 A0A1J1IZA5 A0A151I8Z5 A0A023EWM3 A0A195EDS2 A0A3L8DQ17 A0A195F2T2 A0A151WKN8 A0A158NDG7 A0A232EYR2 T1HAN9 A0A0J7L3P1 A0A0N0BDI5 K7IMG8 A0A154PLK1 A0A2Y9D0U7 A0A1Y9J0K7 A0A1S4GBB2 A0A182YHT9 A0A1B6M4Q5 Q0H6L5 Q0H6L4 Q0H6L3 Q0H6L6 A0A3R7SM39 B4I9E0 A0A336LLG9 B4GTL7 A0A0L0CN79 A0A0K2U468 B4JNT3 A0A2R7VZ47 U4TXE0 N6T9R7 E9J760 A0A0P5FZQ6 A0A182JSF9 A0A0P5JSG3 A0A2P2HW79 A0A0P5ENG7 A0A0P5FXU3 A0A0P6DMC4 A0A0P6G415 A0A0N7ZZC8 A0A0N8ATV5 A0A0P6E8R3 A0A0P6FFV1 A0A0N8DZ12 A0A0P5FWN8 A0A0P5JV90 A0A0P5WVF2 A0A0P4Y595 A0A0P5L9E6 A0A0P5W5G9 A0A0P5USX1 A0A0N8D0C2 A0A0P5JW72 A0A0P6GAG5 A0A0P6GCT4 A0A0P6EHX6 A0A0P5H8R7
EC Number
3.2.1.1
Pubmed
22118469
28756777
26354079
26227816
28004739
18362917
+ More
19820115 25315136 24495485 25401762 17994087 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10839355 10731137 18419581 10839356 11733059 16949836 17372656 21159966 29403074 25474469 30249741 21347285 28648823 20075255 12364791 25244985 16547109 26108605 23537049 21282665
19820115 25315136 24495485 25401762 17994087 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10839355 10731137 18419581 10839356 11733059 16949836 17372656 21159966 29403074 25474469 30249741 21347285 28648823 20075255 12364791 25244985 16547109 26108605 23537049 21282665
EMBL
AGBW02013836
OWR42686.1
RSAL01000117
RVE46820.1
KZ150275
PZC71653.1
+ More
KQ458981 KPJ04455.1 KQ459995 KPJ18506.1 JTDY01000099 KOB78874.1 ODYU01000533 SOQ35454.1 BT030436 ABO52856.1 GEZM01091393 JAV56857.1 KQ971312 EEZ98500.1 GAMC01012561 JAB93994.1 GBHO01038004 JAG05600.1 CH954183 EDV45474.2 GECU01022071 JAS85635.1 CCAG010008295 CH379063 KRT06091.1 KRT06092.1 EAL32502.2 CM000162 EDX00897.2 AE014298 AFH07176.1 AL031128 EU523737 CAA20006.1 CP012528 ALC48213.1 OUUW01000011 SPP86280.1 JXJN01003232 JXJN01003233 JXJN01003234 JXJN01003235 JXJN01003236 CH933810 EDW07358.2 PYGN01000570 PSN44063.1 CH963925 EDW78244.2 CH940660 EDW58073.1 GEDC01012133 JAS25165.1 KQ414590 KOC70405.1 CVRI01000064 CRL05611.1 KQ978323 KYM95138.1 GBBI01004977 JAC13735.1 KQ979039 KYN23256.1 QOIP01000005 RLU22525.1 KQ981855 KYN34885.1 KQ983002 KYQ48406.1 ADTU01001025 ADTU01001026 ADTU01001027 ADTU01001028 ADTU01001029 ADTU01001030 NNAY01001605 OXU23449.1 ACPB03014737 LBMM01000892 KMQ97266.1 KQ435876 KOX70141.1 KQ434960 KZC12637.1 APCN01000822 APCN01000823 AAAB01008987 GEBQ01009071 JAT30906.1 DQ185012 ABB00666.1 DQ185013 ABB00667.1 DQ185014 ABB00668.1 DQ185011 ABB00665.1 QCYY01002917 ROT66601.1 CH480825 EDW43821.1 UFQT01000054 SSX19074.1 CH479190 EDW25887.1 JRES01000262 KNC32874.1 HACA01015637 CDW32998.1 CH916371 EDV92376.1 KK854183 PTY12742.1 KB631698 ERL85462.1 APGK01038521 APGK01038522 KB740960 ENN76999.1 GL768421 EFZ11354.1 GDIQ01269612 JAJ82112.1 GDIQ01200436 JAK51289.1 IACF01000234 LAB66031.1 GDIQ01269613 JAJ82111.1 GDIQ01248869 JAK02856.1 GDIQ01075475 JAN19262.1 GDIQ01041193 JAN53544.1 GDIP01197713 JAJ25689.1 GDIQ01243419 JAK08306.1 GDIQ01077973 JAN16764.1 GDIQ01060483 JAN34254.1 GDIQ01077972 JAN16765.1 GDIQ01250369 JAK01356.1 GDIQ01219125 JAK32600.1 GDIP01081272 JAM22443.1 GDIP01234606 JAI88795.1 GDIQ01173138 JAK78587.1 GDIP01090970 JAM12745.1 GDIP01130465 GDIP01112120 JAL91594.1 GDIP01081271 JAM22444.1 GDIQ01194333 JAK57392.1 GDIQ01047542 JAN47195.1 GDIQ01035143 JAN59594.1 GDIQ01062433 JAN32304.1 GDIQ01236522 JAK15203.1
KQ458981 KPJ04455.1 KQ459995 KPJ18506.1 JTDY01000099 KOB78874.1 ODYU01000533 SOQ35454.1 BT030436 ABO52856.1 GEZM01091393 JAV56857.1 KQ971312 EEZ98500.1 GAMC01012561 JAB93994.1 GBHO01038004 JAG05600.1 CH954183 EDV45474.2 GECU01022071 JAS85635.1 CCAG010008295 CH379063 KRT06091.1 KRT06092.1 EAL32502.2 CM000162 EDX00897.2 AE014298 AFH07176.1 AL031128 EU523737 CAA20006.1 CP012528 ALC48213.1 OUUW01000011 SPP86280.1 JXJN01003232 JXJN01003233 JXJN01003234 JXJN01003235 JXJN01003236 CH933810 EDW07358.2 PYGN01000570 PSN44063.1 CH963925 EDW78244.2 CH940660 EDW58073.1 GEDC01012133 JAS25165.1 KQ414590 KOC70405.1 CVRI01000064 CRL05611.1 KQ978323 KYM95138.1 GBBI01004977 JAC13735.1 KQ979039 KYN23256.1 QOIP01000005 RLU22525.1 KQ981855 KYN34885.1 KQ983002 KYQ48406.1 ADTU01001025 ADTU01001026 ADTU01001027 ADTU01001028 ADTU01001029 ADTU01001030 NNAY01001605 OXU23449.1 ACPB03014737 LBMM01000892 KMQ97266.1 KQ435876 KOX70141.1 KQ434960 KZC12637.1 APCN01000822 APCN01000823 AAAB01008987 GEBQ01009071 JAT30906.1 DQ185012 ABB00666.1 DQ185013 ABB00667.1 DQ185014 ABB00668.1 DQ185011 ABB00665.1 QCYY01002917 ROT66601.1 CH480825 EDW43821.1 UFQT01000054 SSX19074.1 CH479190 EDW25887.1 JRES01000262 KNC32874.1 HACA01015637 CDW32998.1 CH916371 EDV92376.1 KK854183 PTY12742.1 KB631698 ERL85462.1 APGK01038521 APGK01038522 KB740960 ENN76999.1 GL768421 EFZ11354.1 GDIQ01269612 JAJ82112.1 GDIQ01200436 JAK51289.1 IACF01000234 LAB66031.1 GDIQ01269613 JAJ82111.1 GDIQ01248869 JAK02856.1 GDIQ01075475 JAN19262.1 GDIQ01041193 JAN53544.1 GDIP01197713 JAJ25689.1 GDIQ01243419 JAK08306.1 GDIQ01077973 JAN16764.1 GDIQ01060483 JAN34254.1 GDIQ01077972 JAN16765.1 GDIQ01250369 JAK01356.1 GDIQ01219125 JAK32600.1 GDIP01081272 JAM22443.1 GDIP01234606 JAI88795.1 GDIQ01173138 JAK78587.1 GDIP01090970 JAM12745.1 GDIP01130465 GDIP01112120 JAL91594.1 GDIP01081271 JAM22444.1 GDIQ01194333 JAK57392.1 GDIQ01047542 JAN47195.1 GDIQ01035143 JAN59594.1 GDIQ01062433 JAN32304.1 GDIQ01236522 JAK15203.1
Proteomes
UP000007151
UP000283053
UP000053268
UP000053240
UP000037510
UP000007266
+ More
UP000095301 UP000091820 UP000008711 UP000092444 UP000001819 UP000002282 UP000192221 UP000092443 UP000000803 UP000092553 UP000092445 UP000268350 UP000092460 UP000078200 UP000009192 UP000245037 UP000007798 UP000095300 UP000008792 UP000053825 UP000005203 UP000183832 UP000078542 UP000078492 UP000279307 UP000078541 UP000075809 UP000005205 UP000215335 UP000015103 UP000036403 UP000053105 UP000002358 UP000076502 UP000075840 UP000076407 UP000076408 UP000283509 UP000001292 UP000008744 UP000037069 UP000001070 UP000030742 UP000019118 UP000075881
UP000095301 UP000091820 UP000008711 UP000092444 UP000001819 UP000002282 UP000192221 UP000092443 UP000000803 UP000092553 UP000092445 UP000268350 UP000092460 UP000078200 UP000009192 UP000245037 UP000007798 UP000095300 UP000008792 UP000053825 UP000005203 UP000183832 UP000078542 UP000078492 UP000279307 UP000078541 UP000075809 UP000005205 UP000215335 UP000015103 UP000036403 UP000053105 UP000002358 UP000076502 UP000075840 UP000076407 UP000076408 UP000283509 UP000001292 UP000008744 UP000037069 UP000001070 UP000030742 UP000019118 UP000075881
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
A0A212EMI4
A0A3S2TIC9
A0A2W1BJ16
A0A194QHP0
A0A194RR39
A0A0L7LU20
+ More
A0A2H1V5A6 A4IJ78 A0A1Y1KDM4 D6W9J1 A0A1I8N0Y2 W8BXP6 A0A1A9WCP8 A0A0A9WLA3 B3P9H8 A0A1B6IFC7 A0A1B0G446 A0A0R3NYE5 A0A0R3NYW6 Q29J01 B4PXP4 A0A1W4VS68 A0A1A9XAH7 M9NGG5 Q9W596 A0A0M3QYS9 A0A1A9ZRR8 A0A3B0KDE7 A0A1B0ATD1 A0A1A9UPH8 B4L480 A0A2P8YIH9 B4N1K5 A0A1I8PFQ4 B4MD01 A0A1B6DHQ9 A0A0L7RHR1 A0A088ACQ6 A0A1J1IZA5 A0A151I8Z5 A0A023EWM3 A0A195EDS2 A0A3L8DQ17 A0A195F2T2 A0A151WKN8 A0A158NDG7 A0A232EYR2 T1HAN9 A0A0J7L3P1 A0A0N0BDI5 K7IMG8 A0A154PLK1 A0A2Y9D0U7 A0A1Y9J0K7 A0A1S4GBB2 A0A182YHT9 A0A1B6M4Q5 Q0H6L5 Q0H6L4 Q0H6L3 Q0H6L6 A0A3R7SM39 B4I9E0 A0A336LLG9 B4GTL7 A0A0L0CN79 A0A0K2U468 B4JNT3 A0A2R7VZ47 U4TXE0 N6T9R7 E9J760 A0A0P5FZQ6 A0A182JSF9 A0A0P5JSG3 A0A2P2HW79 A0A0P5ENG7 A0A0P5FXU3 A0A0P6DMC4 A0A0P6G415 A0A0N7ZZC8 A0A0N8ATV5 A0A0P6E8R3 A0A0P6FFV1 A0A0N8DZ12 A0A0P5FWN8 A0A0P5JV90 A0A0P5WVF2 A0A0P4Y595 A0A0P5L9E6 A0A0P5W5G9 A0A0P5USX1 A0A0N8D0C2 A0A0P5JW72 A0A0P6GAG5 A0A0P6GCT4 A0A0P6EHX6 A0A0P5H8R7
A0A2H1V5A6 A4IJ78 A0A1Y1KDM4 D6W9J1 A0A1I8N0Y2 W8BXP6 A0A1A9WCP8 A0A0A9WLA3 B3P9H8 A0A1B6IFC7 A0A1B0G446 A0A0R3NYE5 A0A0R3NYW6 Q29J01 B4PXP4 A0A1W4VS68 A0A1A9XAH7 M9NGG5 Q9W596 A0A0M3QYS9 A0A1A9ZRR8 A0A3B0KDE7 A0A1B0ATD1 A0A1A9UPH8 B4L480 A0A2P8YIH9 B4N1K5 A0A1I8PFQ4 B4MD01 A0A1B6DHQ9 A0A0L7RHR1 A0A088ACQ6 A0A1J1IZA5 A0A151I8Z5 A0A023EWM3 A0A195EDS2 A0A3L8DQ17 A0A195F2T2 A0A151WKN8 A0A158NDG7 A0A232EYR2 T1HAN9 A0A0J7L3P1 A0A0N0BDI5 K7IMG8 A0A154PLK1 A0A2Y9D0U7 A0A1Y9J0K7 A0A1S4GBB2 A0A182YHT9 A0A1B6M4Q5 Q0H6L5 Q0H6L4 Q0H6L3 Q0H6L6 A0A3R7SM39 B4I9E0 A0A336LLG9 B4GTL7 A0A0L0CN79 A0A0K2U468 B4JNT3 A0A2R7VZ47 U4TXE0 N6T9R7 E9J760 A0A0P5FZQ6 A0A182JSF9 A0A0P5JSG3 A0A2P2HW79 A0A0P5ENG7 A0A0P5FXU3 A0A0P6DMC4 A0A0P6G415 A0A0N7ZZC8 A0A0N8ATV5 A0A0P6E8R3 A0A0P6FFV1 A0A0N8DZ12 A0A0P5FWN8 A0A0P5JV90 A0A0P5WVF2 A0A0P4Y595 A0A0P5L9E6 A0A0P5W5G9 A0A0P5USX1 A0A0N8D0C2 A0A0P5JW72 A0A0P6GAG5 A0A0P6GCT4 A0A0P6EHX6 A0A0P5H8R7
Ontologies
GO
GO:0005874
GO:0000226
GO:0008017
GO:0030425
GO:0042995
GO:0003779
GO:0005829
GO:0015631
GO:0007409
GO:0005875
GO:0045202
GO:0031114
GO:0043025
GO:0016358
GO:0008582
GO:0008088
GO:0030424
GO:0019900
GO:0008355
GO:0043524
GO:0048813
GO:0060052
GO:0048749
GO:1901215
GO:0007528
GO:0007017
GO:0015630
GO:0007399
GO:0103025
GO:0005975
GO:0043169
GO:0004556
PANTHER
Topology
Subcellular location
Cytoplasm
Cytoskeleton
Cytoskeleton
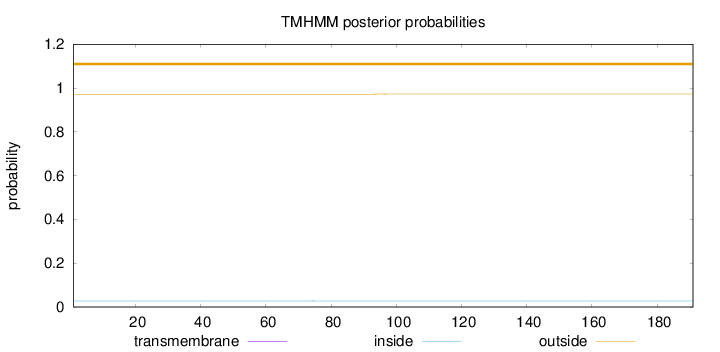
Length:
191
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0143
Exp number, first 60 AAs:
0.00038
Total prob of N-in:
0.02780
outside
1 - 191
Population Genetic Test Statistics
Pi
22.861197
Theta
20.723021
Tajima's D
-1.223354
CLR
0.780051
CSRT
0.101694915254237
Interpretation
Uncertain
