Gene
KWMTBOMO12778
Pre Gene Modal
BGIBMGA013095
Annotation
reverse_transcriptase_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 2.434
Sequence
CDS
ATGGCTCAACTTATAGGAAACATATCTGAGTTTGATTATAAAACCCAAGAATGGCAGATCTTTTACAAGAAATTAGAAAATTTCCTAAAGTTGAACAAAGTTTTAGCTGAAGATTGGTGCTCCGTGCTTATTGCACATCTTTCAGATGATTCGTATCGGCTTGTGAGGAACCTCGTACATCCTAAGGAGGTAGAAATAGTACAATGGACTGAGCTGGTTAGAGTACTCAACGGACATTTTCAACCGAAAAGATCGACTTTTGCCGACAGGGCAAAATTTTATGGTGCGATCAGAATGGACGGTGAGAAAGTCGAAGATTGGGCGGCACGTCTGAGGGGACTCGCCGTGCACTGTAATTTCGGTACAGAATTAGATACGTTGCTACGTGATCGATTTATTCTAGGCCTGAAAGTAGGTCCAGAAAGAGATCGATTGTTCGAACAAGACTCGAACTCCCTAACGCTTTCGAGAGCTATTGAAATAGCCCAGGAGACTGCGTGTGCGCGGGCGGCGAGAGCTGATGTTGATGGTAGTGAGCAGCTTATCAAACAAGAGATGGTGTATCGTGTCAGCGAGCGTCGGGCGCGGGGCGCCTCGGCGCAGGCGGAGTCGTCGTGGGCATCGGCTGCAGCCGCGCGCTGTGCAGTGTGCGGGTTGAAAGGTCATTCCGTAGAAAAGTGCCGTTTCAAAGACTATAAGTGTATGATTTGTGGTAAAAAGGGTCACCTTAAGAAAGTGTGCAGTGTCAAAAAGGTCGGAAAACTGTCGTTAAATAATATGGATGATAACGAAAGTGATGGACACGAATGTAAAAAATGTGATTTGTATAATTTAAGGTATGTTCGCTACGATCCTATTAAGTTGAACATTTCTATAAATAATAAGTTTTATTCTATGGAATTAGATTGCGGAGCTGCTCGTAGTGTAATTTCAGAAAGATTTTACAGGGATAACTTCCCATTATACAAATTAATTAGAGCGGAATACCAAGAGGTGCAGCAGATCTTGAATGAATTCCCTGACATTTGGAGAGATGAATTGGGCCGTTTTAATAAATATGAAGTGCATTTACACTTGAAAGAAAATACAACACCTAAATTTTTTAAGCCACGCCAAATACCGTTTGCTCTAAAGGACAAGGTTGAAGAAGAATTACATAGGCTTGTCGAAGCAGGTATATTAGTACCAGTACAGTTTTCAGATTATGCTACGCCCATAGTACCAGTTCTGAAGGAGAATGGTAAAGTGAAAATAGCTGGAGATTTTTCCTGTACCCTAAATAAAGTTTTAAAAATTGACAAATATCCATTACCCCACATAGACGAGGTTTTCGCGAAGTTAGGCGGCGGCGAGCATTATTCTAAGCTAGATTTAAGTAATGCCTATAATCAAATTTGTTTGTCTCAAGCCTCTCAAGACCTCACAACTATAAATACACCGAAAGGACTTTTCAAATATACACGCTTAGTGTACGGTCTCGCGAATGCCCCAGCTATTTTCCAGCGCACAATAGAATCTCTAGTAGTAGGTATTGAAGGAGTGTCGTGTTGGAATTTTATTCCTAATGCATCCTCATTATTAAGTCCGCTGAACGAATTATTAAAAGACGACATAAAATGGCACTGGGATAAGCGACAGCAGCTTGCGTTTGATTCCGTGCGAAAAGAACTCGCCTCGGACCGCGTACTATCACATTTCGATCCGAGGGCGCAGCTAACATTGACAGTGGATGCGGGCCCGTCTGGTCTCGGCGCGGGGGAGGATGAGGAGGGCGATGAATGGGAGGAGGCTAGTGAAAACATCGCAGCTGCCAGCCTGCCGACCGAGTGTGAACCTGATTACCCTATTGTAGCATCAGATGACCCCGATAACGTGAGATTAGAAGAGAGGGCGCCAGATAATGAAAGACAGTGTGGTCGACCAAAGAGGACAAGAAAAGTAGTAGATTATAAGAAGTATTTTTAA
Protein
MAQLIGNISEFDYKTQEWQIFYKKLENFLKLNKVLAEDWCSVLIAHLSDDSYRLVRNLVHPKEVEIVQWTELVRVLNGHFQPKRSTFADRAKFYGAIRMDGEKVEDWAARLRGLAVHCNFGTELDTLLRDRFILGLKVGPERDRLFEQDSNSLTLSRAIEIAQETACARAARADVDGSEQLIKQEMVYRVSERRARGASAQAESSWASAAAARCAVCGLKGHSVEKCRFKDYKCMICGKKGHLKKVCSVKKVGKLSLNNMDDNESDGHECKKCDLYNLRYVRYDPIKLNISINNKFYSMELDCGAARSVISERFYRDNFPLYKLIRAEYQEVQQILNEFPDIWRDELGRFNKYEVHLHLKENTTPKFFKPRQIPFALKDKVEEELHRLVEAGILVPVQFSDYATPIVPVLKENGKVKIAGDFSCTLNKVLKIDKYPLPHIDEVFAKLGGGEHYSKLDLSNAYNQICLSQASQDLTTINTPKGLFKYTRLVYGLANAPAIFQRTIESLVVGIEGVSCWNFIPNASSLLSPLNELLKDDIKWHWDKRQQLAFDSVRKELASDRVLSHFDPRAQLTLTVDAGPSGLGAGEDEEGDEWEEASENIAAASLPTECEPDYPIVASDDPDNVRLEERAPDNERQCGRPKRTRKVVDYKKYF
Summary
Uniprot
X1XT86
X1WTX7
Q5KTM2
J9KFN3
A0A1Y1KCE7
J9LDN7
+ More
A0A2B4S417 J9KDA1 A0A3P8QD94 A0A3P8NHI5 J9LBK5 A0A1Y1NFA4 A0A3P8RGT3 A0A0A9WT47 J9L142 A0A3B3R045 A0A2S2PU23 A0A1B6DD07 A0A1B6CPB9 A0A0J7KGA3 A0A3S0ZSV8 A0A293M1Z2 A0A085LXZ4 A0A1Y1M058 A0A1S4EQA3 A0A1X7UD72 A0A2S2N957 A0A147BQ83 X1WRQ5 H3AL86 A0A2B4T0D2 B7QBI4 A0A0V0TT39 A0A090XAL0 A0A0V0ZE23 A0A0V0ZDL9 A0A267G4K7 A0A267E8T4 A0A267EF28
A0A2B4S417 J9KDA1 A0A3P8QD94 A0A3P8NHI5 J9LBK5 A0A1Y1NFA4 A0A3P8RGT3 A0A0A9WT47 J9L142 A0A3B3R045 A0A2S2PU23 A0A1B6DD07 A0A1B6CPB9 A0A0J7KGA3 A0A3S0ZSV8 A0A293M1Z2 A0A085LXZ4 A0A1Y1M058 A0A1S4EQA3 A0A1X7UD72 A0A2S2N957 A0A147BQ83 X1WRQ5 H3AL86 A0A2B4T0D2 B7QBI4 A0A0V0TT39 A0A090XAL0 A0A0V0ZE23 A0A0V0ZDL9 A0A267G4K7 A0A267E8T4 A0A267EF28
EMBL
ABLF02012266
ABLF02012271
ABLF02008248
ABLF02008249
ABLF02008250
ABLF02035153
+ More
ABLF02037855 ABLF02037866 ABLF02061872 AB126055 BAD86655.1 ABLF02036937 GEZM01092768 GEZM01092767 JAV56467.1 ABLF02030002 LSMT01000205 PFX23520.1 ABLF02025265 ABLF02025271 ABLF02025272 ABLF02038551 ABLF02038554 ABLF02042419 GEZM01009753 JAV94337.1 GBHO01031997 JAG11607.1 ABLF02012827 ABLF02012828 ABLF02015552 ABLF02041322 GGMR01020219 MBY32838.1 GEDC01013717 JAS23581.1 GEDC01021971 JAS15327.1 LBMM01007751 KMQ89453.1 RQTK01000185 RUS85038.1 GFWV01009597 MAA34326.1 KL363263 KFD49840.1 GEZM01043158 JAV79264.1 GGMR01001051 MBY13670.1 GEGO01002490 JAR92914.1 ABLF02024359 ABLF02057220 AFYH01176858 LSMT01000001 PFX35006.1 ABJB010082761 DS901637 EEC16206.1 JYDJ01000151 KRX42156.1 GBIH01000758 JAC93952.1 JYDQ01000223 KRY10622.1 KRY10623.1 NIVC01000594 PAA80182.1 NIVC01002515 PAA57229.1 NIVC01002188 PAA60150.1
ABLF02037855 ABLF02037866 ABLF02061872 AB126055 BAD86655.1 ABLF02036937 GEZM01092768 GEZM01092767 JAV56467.1 ABLF02030002 LSMT01000205 PFX23520.1 ABLF02025265 ABLF02025271 ABLF02025272 ABLF02038551 ABLF02038554 ABLF02042419 GEZM01009753 JAV94337.1 GBHO01031997 JAG11607.1 ABLF02012827 ABLF02012828 ABLF02015552 ABLF02041322 GGMR01020219 MBY32838.1 GEDC01013717 JAS23581.1 GEDC01021971 JAS15327.1 LBMM01007751 KMQ89453.1 RQTK01000185 RUS85038.1 GFWV01009597 MAA34326.1 KL363263 KFD49840.1 GEZM01043158 JAV79264.1 GGMR01001051 MBY13670.1 GEGO01002490 JAR92914.1 ABLF02024359 ABLF02057220 AFYH01176858 LSMT01000001 PFX35006.1 ABJB010082761 DS901637 EEC16206.1 JYDJ01000151 KRX42156.1 GBIH01000758 JAC93952.1 JYDQ01000223 KRY10622.1 KRY10623.1 NIVC01000594 PAA80182.1 NIVC01002515 PAA57229.1 NIVC01002188 PAA60150.1
Proteomes
PRIDE
Pfam
Interpro
IPR021109
Peptidase_aspartic_dom_sf
+ More
IPR041577 RT_RNaseH_2
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR034128 K02A2.6-like
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR041373 RT_RNaseH
IPR006612 THAP_Znf
IPR001969 Aspartic_peptidase_AS
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR001995 Peptidase_A2_cat
IPR018061 Retropepsins
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR036116 FN3_sf
IPR005312 DUF1759
IPR012341 6hp_glycosidase-like_sf
IPR020464 LanC-like_prot_euk
IPR007822 LANC-like
IPR041577 RT_RNaseH_2
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR034128 K02A2.6-like
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR041373 RT_RNaseH
IPR006612 THAP_Znf
IPR001969 Aspartic_peptidase_AS
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR001995 Peptidase_A2_cat
IPR018061 Retropepsins
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR036116 FN3_sf
IPR005312 DUF1759
IPR012341 6hp_glycosidase-like_sf
IPR020464 LanC-like_prot_euk
IPR007822 LANC-like
SUPFAM
Gene 3D
ProteinModelPortal
X1XT86
X1WTX7
Q5KTM2
J9KFN3
A0A1Y1KCE7
J9LDN7
+ More
A0A2B4S417 J9KDA1 A0A3P8QD94 A0A3P8NHI5 J9LBK5 A0A1Y1NFA4 A0A3P8RGT3 A0A0A9WT47 J9L142 A0A3B3R045 A0A2S2PU23 A0A1B6DD07 A0A1B6CPB9 A0A0J7KGA3 A0A3S0ZSV8 A0A293M1Z2 A0A085LXZ4 A0A1Y1M058 A0A1S4EQA3 A0A1X7UD72 A0A2S2N957 A0A147BQ83 X1WRQ5 H3AL86 A0A2B4T0D2 B7QBI4 A0A0V0TT39 A0A090XAL0 A0A0V0ZE23 A0A0V0ZDL9 A0A267G4K7 A0A267E8T4 A0A267EF28
A0A2B4S417 J9KDA1 A0A3P8QD94 A0A3P8NHI5 J9LBK5 A0A1Y1NFA4 A0A3P8RGT3 A0A0A9WT47 J9L142 A0A3B3R045 A0A2S2PU23 A0A1B6DD07 A0A1B6CPB9 A0A0J7KGA3 A0A3S0ZSV8 A0A293M1Z2 A0A085LXZ4 A0A1Y1M058 A0A1S4EQA3 A0A1X7UD72 A0A2S2N957 A0A147BQ83 X1WRQ5 H3AL86 A0A2B4T0D2 B7QBI4 A0A0V0TT39 A0A090XAL0 A0A0V0ZE23 A0A0V0ZDL9 A0A267G4K7 A0A267E8T4 A0A267EF28
PDB
4OL8
E-value=2.20389e-16,
Score=211
Ontologies
KEGG
GO
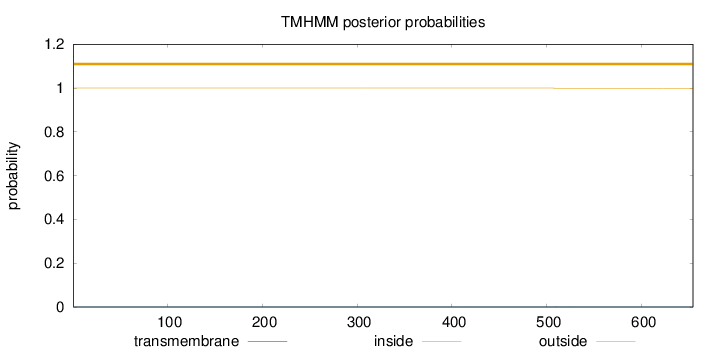
Topology
Length:
654
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00322
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00029
outside
1 - 654
Population Genetic Test Statistics
Pi
11.035767
Theta
12.01733
Tajima's D
-0.646562
CLR
3.65391
CSRT
0.214689265536723
Interpretation
Uncertain
