Gene
KWMTBOMO12776
Pre Gene Modal
BGIBMGA007152
Annotation
PREDICTED:_DDB1-_and_CUL4-associated_factor_12_[Bombyx_mori]
Full name
Deoxyhypusine hydroxylase
Alternative Name
Deoxyhypusine dioxygenase
Deoxyhypusine monooxygenase
Deoxyhypusine monooxygenase
Location in the cell
Nuclear Reliability : 2.454
Sequence
CDS
ATGGCGCAGACTCTTCGTAGAGCAGTAGAGGGCAGGTACCCCTCTTGCTATTTACCCAATCGATTGAAAGAAAGGAAAGCCAAAGCTAAAGCTTTGAGATTAGAGCAACGTAAACCAGACAAACCGGAAGATTTTGTTTGCTATTCAGACAGTGACAGTGAAGAGGAAACACCGACGCAACAACATCAAATTTTATCAACGTCATATAATTTTGTCGACTATGTACGTTCAAGAGAAATACTTGCTCCTGAGCCTCGATCAGTCGACCCATCGTATGCAACCCGACACATGCTCACACATGATATGTTTCATGAGACACCAGTCAACCTCGGTAACATGAACAAGGTCTTCTGCTCACAATGGCTATCAGACAGACAAGTTGTATTTGGCACCAAATGTAACAAGTTAATGGTGTATGATGTCCGCACTCGGTCCTTGGACGCCATCCCCACACTGCGCCCGCGCGCCGCCTGGCCGCCGCCGGACCACCAGACCGGCATACACGCGCTCCAGATCAATCCTTCCAGAACACTGCTAGCGACCGGTGCGAGGAATTCTTGTGAATTAGCCATATACAGCCTACCTACACTAGACCCGGTCTGCGTTGGAGAAGCTCATAAAGATTGGATCCTGGACATGTGCTGGTTGGACGACGAGTTCGTGGTGACGGGGTCGCGCGATAATAAGCTTGCACTGTGGCGCGCCCCGGCCGCCCCCCCGCCCCGCGCGCACCTGCACCACTACGTGCACCCCCACGCCGTGCGGGAGTGTCGGTCGGGTCTAAAAGTCCGAGCGCTGACGTTCAACGAGCGCTGGCGGGAGATAGCGGCACTTACCCTCAACGGCTACATACACGTGTTCAACGCGGAGACCTTCCGCCAGACGCTGTCCAGGAAGCTGCCCTCGTGCCAGGACTTGGTGTGTCTAGCCACCCAGCAAGAGAGCGGCATGTACGCGATCGGCTGCAAGTCGTACACGCTGCTGCTGGACTGCCGCACGCTGCAGTCCGTCAAGAAGGTCACCTCCCGGTACTCCGGCTGCGGGGTCCGCTCCGCCAGCTTCCGGGGGAACATGCTCACCATCGGGACCGGACTGGGTCTCCTGATGTTCTACGACCTGCGCGCGGGTAAGTACCTGGAGAGCAGCATCCACTCGTCCAAGACGGTGACGCTGAAGGCGTCCCGCGGCTACCTGTTCCCCGAGGAGGAGGCGGAGGGCGGCGTCAAGCACGTGCCCGCCATCTACACGCACTGCTACGACTCCTCGGGCACCAGGGTGTTCACCGCGGGCGGGCCCCTACCGGCTCCCCTAGTCGGGAACTACGCAGCCATATGGCAGTAG
Protein
MAQTLRRAVEGRYPSCYLPNRLKERKAKAKALRLEQRKPDKPEDFVCYSDSDSEEETPTQQHQILSTSYNFVDYVRSREILAPEPRSVDPSYATRHMLTHDMFHETPVNLGNMNKVFCSQWLSDRQVVFGTKCNKLMVYDVRTRSLDAIPTLRPRAAWPPPDHQTGIHALQINPSRTLLATGARNSCELAIYSLPTLDPVCVGEAHKDWILDMCWLDDEFVVTGSRDNKLALWRAPAAPPPRAHLHHYVHPHAVRECRSGLKVRALTFNERWREIAALTLNGYIHVFNAETFRQTLSRKLPSCQDLVCLATQQESGMYAIGCKSYTLLLDCRTLQSVKKVTSRYSGCGVRSASFRGNMLTIGTGLGLLMFYDLRAGKYLESSIHSSKTVTLKASRGYLFPEEEAEGGVKHVPAIYTHCYDSSGTRVFTAGGPLPAPLVGNYAAIWQ
Summary
Description
Catalyzes the hydroxylation of the N(6)-(4-aminobutyl)-L-lysine intermediate to form hypusine, an essential post-translational modification only found in mature eIF-5A factor.
Catalytic Activity
[eIF5A protein]-deoxyhypusine + AH2 + O2 = [eIF5A protein]-hypusine + A + H2O
Cofactor
Fe(2+)
Similarity
Belongs to the deoxyhypusine hydroxylase family.
Uniprot
H9JCA7
A0A2H1WL49
A0A194QG40
B0X2Z8
A0A1L8DNM1
A0A182PM82
+ More
A0A182RH58 A0A182JYZ8 A0A182ND54 A0A1Q3F3B3 A0A182JLA1 A0A182T2Z8 A0A084VWL7 Q17DJ1 A0A182UBS0 A0A182X3A9 A0A182MJR0 A0A1W4WT74 A0A182VQ15 A0A182HZF0 Q7QKC2 A0A182LQG2 A0A182VKJ3 A0A2P8YZY5 A0A182QF97 A0A1B0C9L4 A0A1J1IL38 A0A1Y1LFN9 A0A2J7QK27 D6WCT6 A0A336MNM5 A0A067RUN9 A0A2M4APH4 W5J4K4 A0A182F6R1 A0A2M4BM77 A0A2M3Z9C2 A0A195EN51 F4WRW0 A0A195BUX7 A0A158NEK5 A0A151XIU2 A0A195FLA6 A0A195BXW5 E2AFP2 A0A026WYK8 E9ID87 E2B5Y4 T1HMI4 A0A0V0GB69 A0A182Y4R5 A0A1B6L9I8 A0A0A9W184 A0A1I8PTU3 A0A0L7RFL8 A0A2A3EPQ9 A0A088AKW5 A0A1B6JL94 A0A154P0N9 A0A1A9YG53 A0A1B6FEX8 K7JAI9 A0A1A9VBC6 A0A1B0AK90 A0A1B6DPS3 A0A3B0JQM3 A0A1W4UBC4 B4NH58 B4PN20 B4K5L6 B4HHI2 Q9VGE3 B3M032 A0A1I8MU32 Q295S9 B4GFQ6 B4LWG4 B4JT78 B3P1H5 N6TRP9 A0A0M9A5C4 A0A0K8VQ02 A0A034VCG9 A0A1A9WJC9 W8BEG9 A0A1B0F9U7 N6T871 A0A310SH56 A0A2W1BN22 A0A212F9W7 E0VJR3 A0A2S2NJR9 X1WTX5 A0A023EQ92 A0A0M3QYI5 B4QTH9 A0A232FBQ3
A0A182RH58 A0A182JYZ8 A0A182ND54 A0A1Q3F3B3 A0A182JLA1 A0A182T2Z8 A0A084VWL7 Q17DJ1 A0A182UBS0 A0A182X3A9 A0A182MJR0 A0A1W4WT74 A0A182VQ15 A0A182HZF0 Q7QKC2 A0A182LQG2 A0A182VKJ3 A0A2P8YZY5 A0A182QF97 A0A1B0C9L4 A0A1J1IL38 A0A1Y1LFN9 A0A2J7QK27 D6WCT6 A0A336MNM5 A0A067RUN9 A0A2M4APH4 W5J4K4 A0A182F6R1 A0A2M4BM77 A0A2M3Z9C2 A0A195EN51 F4WRW0 A0A195BUX7 A0A158NEK5 A0A151XIU2 A0A195FLA6 A0A195BXW5 E2AFP2 A0A026WYK8 E9ID87 E2B5Y4 T1HMI4 A0A0V0GB69 A0A182Y4R5 A0A1B6L9I8 A0A0A9W184 A0A1I8PTU3 A0A0L7RFL8 A0A2A3EPQ9 A0A088AKW5 A0A1B6JL94 A0A154P0N9 A0A1A9YG53 A0A1B6FEX8 K7JAI9 A0A1A9VBC6 A0A1B0AK90 A0A1B6DPS3 A0A3B0JQM3 A0A1W4UBC4 B4NH58 B4PN20 B4K5L6 B4HHI2 Q9VGE3 B3M032 A0A1I8MU32 Q295S9 B4GFQ6 B4LWG4 B4JT78 B3P1H5 N6TRP9 A0A0M9A5C4 A0A0K8VQ02 A0A034VCG9 A0A1A9WJC9 W8BEG9 A0A1B0F9U7 N6T871 A0A310SH56 A0A2W1BN22 A0A212F9W7 E0VJR3 A0A2S2NJR9 X1WTX5 A0A023EQ92 A0A0M3QYI5 B4QTH9 A0A232FBQ3
EC Number
1.14.99.29
Pubmed
19121390
26354079
24438588
17510324
12364791
14747013
+ More
17210077 20966253 29403074 28004739 18362917 19820115 24845553 20920257 23761445 21719571 21347285 20798317 24508170 30249741 21282665 25244985 25401762 26823975 20075255 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 15632085 23537049 25348373 24495485 28756777 22118469 20566863 24945155 28648823
17210077 20966253 29403074 28004739 18362917 19820115 24845553 20920257 23761445 21719571 21347285 20798317 24508170 30249741 21282665 25244985 25401762 26823975 20075255 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 15632085 23537049 25348373 24495485 28756777 22118469 20566863 24945155 28648823
EMBL
BABH01031615
ODYU01009375
SOQ53758.1
KQ458981
KPJ04457.1
DS232304
+ More
EDS39500.1 GFDF01006144 JAV07940.1 GFDL01013028 JAV22017.1 ATLV01017659 KE525181 KFB42361.1 CH477293 EAT44511.1 AXCM01000816 APCN01001973 AAAB01008799 EAA03757.4 PYGN01000262 PSN49804.1 AXCN02001214 AJWK01002461 CVRI01000049 CRK99177.1 GEZM01057108 JAV72443.1 NEVH01013278 PNF28940.1 KQ971311 EEZ99060.1 UFQS01001276 UFQT01001276 SSX10085.1 SSX29807.1 KK852405 KDR24525.1 GGFK01009370 MBW42691.1 ADMH02002112 ETN58851.1 GGFJ01004912 MBW54053.1 GGFM01004334 MBW25085.1 KQ978625 KYN29596.1 GL888292 EGI63117.1 KQ976401 KYM92387.1 ADTU01013478 KQ982080 KYQ60326.1 KQ981490 KYN41193.1 KQ978501 KYM93437.1 GL439118 EFN67793.1 KK107064 QOIP01000008 EZA60856.1 RLU19467.1 RLU19624.1 GL762454 EFZ21425.1 GL445887 EFN88953.1 ACPB03015269 GECL01000764 JAP05360.1 GEBQ01030526 GEBQ01019605 JAT09451.1 JAT20372.1 GBHO01043381 GBHO01014904 GDHC01017196 JAG00223.1 JAG28700.1 JAQ01433.1 KQ414599 KOC69772.1 KZ288195 PBC33773.1 GECU01007702 JAT00005.1 KQ434791 KZC05392.1 GECZ01021003 GECZ01011459 GECZ01009193 JAS48766.1 JAS58310.1 JAS60576.1 AAZX01001753 GEDC01009628 JAS27670.1 OUUW01000008 SPP84477.1 CH964272 EDW84555.1 CM000160 EDW97032.1 CH933806 EDW14053.1 CH480815 EDW42521.1 AE014297 AY069488 AAF54740.2 AAL39633.1 AGB95877.1 CH902617 EDV41989.2 CM000070 EAL28629.1 CH479182 EDW34441.1 CH940650 EDW66601.1 CH916373 EDV94968.1 CH954181 EDV49434.1 APGK01006423 APGK01006424 KB736759 KB632379 ENN83144.1 ERL94087.1 KQ435750 KOX76263.1 GDHF01011391 JAI40923.1 GAKP01019731 GAKP01019729 GAKP01019727 JAC39223.1 GAMC01009498 GAMC01009496 JAB97057.1 CCAG010002599 APGK01040417 KB740979 ENN76429.1 KQ764626 OAD54463.1 KZ150013 PZC75024.1 AGBW02009556 OWR50524.1 DS235226 EEB13619.1 GGMR01004814 MBY17433.1 ABLF02036816 ABLF02042531 GAPW01001856 JAC11742.1 CP012526 ALC47679.1 CM000364 EDX13284.1 NNAY01000518 OXU27898.1
EDS39500.1 GFDF01006144 JAV07940.1 GFDL01013028 JAV22017.1 ATLV01017659 KE525181 KFB42361.1 CH477293 EAT44511.1 AXCM01000816 APCN01001973 AAAB01008799 EAA03757.4 PYGN01000262 PSN49804.1 AXCN02001214 AJWK01002461 CVRI01000049 CRK99177.1 GEZM01057108 JAV72443.1 NEVH01013278 PNF28940.1 KQ971311 EEZ99060.1 UFQS01001276 UFQT01001276 SSX10085.1 SSX29807.1 KK852405 KDR24525.1 GGFK01009370 MBW42691.1 ADMH02002112 ETN58851.1 GGFJ01004912 MBW54053.1 GGFM01004334 MBW25085.1 KQ978625 KYN29596.1 GL888292 EGI63117.1 KQ976401 KYM92387.1 ADTU01013478 KQ982080 KYQ60326.1 KQ981490 KYN41193.1 KQ978501 KYM93437.1 GL439118 EFN67793.1 KK107064 QOIP01000008 EZA60856.1 RLU19467.1 RLU19624.1 GL762454 EFZ21425.1 GL445887 EFN88953.1 ACPB03015269 GECL01000764 JAP05360.1 GEBQ01030526 GEBQ01019605 JAT09451.1 JAT20372.1 GBHO01043381 GBHO01014904 GDHC01017196 JAG00223.1 JAG28700.1 JAQ01433.1 KQ414599 KOC69772.1 KZ288195 PBC33773.1 GECU01007702 JAT00005.1 KQ434791 KZC05392.1 GECZ01021003 GECZ01011459 GECZ01009193 JAS48766.1 JAS58310.1 JAS60576.1 AAZX01001753 GEDC01009628 JAS27670.1 OUUW01000008 SPP84477.1 CH964272 EDW84555.1 CM000160 EDW97032.1 CH933806 EDW14053.1 CH480815 EDW42521.1 AE014297 AY069488 AAF54740.2 AAL39633.1 AGB95877.1 CH902617 EDV41989.2 CM000070 EAL28629.1 CH479182 EDW34441.1 CH940650 EDW66601.1 CH916373 EDV94968.1 CH954181 EDV49434.1 APGK01006423 APGK01006424 KB736759 KB632379 ENN83144.1 ERL94087.1 KQ435750 KOX76263.1 GDHF01011391 JAI40923.1 GAKP01019731 GAKP01019729 GAKP01019727 JAC39223.1 GAMC01009498 GAMC01009496 JAB97057.1 CCAG010002599 APGK01040417 KB740979 ENN76429.1 KQ764626 OAD54463.1 KZ150013 PZC75024.1 AGBW02009556 OWR50524.1 DS235226 EEB13619.1 GGMR01004814 MBY17433.1 ABLF02036816 ABLF02042531 GAPW01001856 JAC11742.1 CP012526 ALC47679.1 CM000364 EDX13284.1 NNAY01000518 OXU27898.1
Proteomes
UP000005204
UP000053268
UP000002320
UP000075885
UP000075900
UP000075881
+ More
UP000075884 UP000075880 UP000075901 UP000030765 UP000008820 UP000075902 UP000076407 UP000075883 UP000192223 UP000075920 UP000075840 UP000007062 UP000075882 UP000075903 UP000245037 UP000075886 UP000092461 UP000183832 UP000235965 UP000007266 UP000027135 UP000000673 UP000069272 UP000078492 UP000007755 UP000078540 UP000005205 UP000075809 UP000078541 UP000078542 UP000000311 UP000053097 UP000279307 UP000008237 UP000015103 UP000076408 UP000095300 UP000053825 UP000242457 UP000005203 UP000076502 UP000092443 UP000002358 UP000078200 UP000092445 UP000268350 UP000192221 UP000007798 UP000002282 UP000009192 UP000001292 UP000000803 UP000007801 UP000095301 UP000001819 UP000008744 UP000008792 UP000001070 UP000008711 UP000019118 UP000030742 UP000053105 UP000091820 UP000092444 UP000007151 UP000009046 UP000007819 UP000092553 UP000000304 UP000215335
UP000075884 UP000075880 UP000075901 UP000030765 UP000008820 UP000075902 UP000076407 UP000075883 UP000192223 UP000075920 UP000075840 UP000007062 UP000075882 UP000075903 UP000245037 UP000075886 UP000092461 UP000183832 UP000235965 UP000007266 UP000027135 UP000000673 UP000069272 UP000078492 UP000007755 UP000078540 UP000005205 UP000075809 UP000078541 UP000078542 UP000000311 UP000053097 UP000279307 UP000008237 UP000015103 UP000076408 UP000095300 UP000053825 UP000242457 UP000005203 UP000076502 UP000092443 UP000002358 UP000078200 UP000092445 UP000268350 UP000192221 UP000007798 UP000002282 UP000009192 UP000001292 UP000000803 UP000007801 UP000095301 UP000001819 UP000008744 UP000008792 UP000001070 UP000008711 UP000019118 UP000030742 UP000053105 UP000091820 UP000092444 UP000007151 UP000009046 UP000007819 UP000092553 UP000000304 UP000215335
Interpro
Gene 3D
ProteinModelPortal
H9JCA7
A0A2H1WL49
A0A194QG40
B0X2Z8
A0A1L8DNM1
A0A182PM82
+ More
A0A182RH58 A0A182JYZ8 A0A182ND54 A0A1Q3F3B3 A0A182JLA1 A0A182T2Z8 A0A084VWL7 Q17DJ1 A0A182UBS0 A0A182X3A9 A0A182MJR0 A0A1W4WT74 A0A182VQ15 A0A182HZF0 Q7QKC2 A0A182LQG2 A0A182VKJ3 A0A2P8YZY5 A0A182QF97 A0A1B0C9L4 A0A1J1IL38 A0A1Y1LFN9 A0A2J7QK27 D6WCT6 A0A336MNM5 A0A067RUN9 A0A2M4APH4 W5J4K4 A0A182F6R1 A0A2M4BM77 A0A2M3Z9C2 A0A195EN51 F4WRW0 A0A195BUX7 A0A158NEK5 A0A151XIU2 A0A195FLA6 A0A195BXW5 E2AFP2 A0A026WYK8 E9ID87 E2B5Y4 T1HMI4 A0A0V0GB69 A0A182Y4R5 A0A1B6L9I8 A0A0A9W184 A0A1I8PTU3 A0A0L7RFL8 A0A2A3EPQ9 A0A088AKW5 A0A1B6JL94 A0A154P0N9 A0A1A9YG53 A0A1B6FEX8 K7JAI9 A0A1A9VBC6 A0A1B0AK90 A0A1B6DPS3 A0A3B0JQM3 A0A1W4UBC4 B4NH58 B4PN20 B4K5L6 B4HHI2 Q9VGE3 B3M032 A0A1I8MU32 Q295S9 B4GFQ6 B4LWG4 B4JT78 B3P1H5 N6TRP9 A0A0M9A5C4 A0A0K8VQ02 A0A034VCG9 A0A1A9WJC9 W8BEG9 A0A1B0F9U7 N6T871 A0A310SH56 A0A2W1BN22 A0A212F9W7 E0VJR3 A0A2S2NJR9 X1WTX5 A0A023EQ92 A0A0M3QYI5 B4QTH9 A0A232FBQ3
A0A182RH58 A0A182JYZ8 A0A182ND54 A0A1Q3F3B3 A0A182JLA1 A0A182T2Z8 A0A084VWL7 Q17DJ1 A0A182UBS0 A0A182X3A9 A0A182MJR0 A0A1W4WT74 A0A182VQ15 A0A182HZF0 Q7QKC2 A0A182LQG2 A0A182VKJ3 A0A2P8YZY5 A0A182QF97 A0A1B0C9L4 A0A1J1IL38 A0A1Y1LFN9 A0A2J7QK27 D6WCT6 A0A336MNM5 A0A067RUN9 A0A2M4APH4 W5J4K4 A0A182F6R1 A0A2M4BM77 A0A2M3Z9C2 A0A195EN51 F4WRW0 A0A195BUX7 A0A158NEK5 A0A151XIU2 A0A195FLA6 A0A195BXW5 E2AFP2 A0A026WYK8 E9ID87 E2B5Y4 T1HMI4 A0A0V0GB69 A0A182Y4R5 A0A1B6L9I8 A0A0A9W184 A0A1I8PTU3 A0A0L7RFL8 A0A2A3EPQ9 A0A088AKW5 A0A1B6JL94 A0A154P0N9 A0A1A9YG53 A0A1B6FEX8 K7JAI9 A0A1A9VBC6 A0A1B0AK90 A0A1B6DPS3 A0A3B0JQM3 A0A1W4UBC4 B4NH58 B4PN20 B4K5L6 B4HHI2 Q9VGE3 B3M032 A0A1I8MU32 Q295S9 B4GFQ6 B4LWG4 B4JT78 B3P1H5 N6TRP9 A0A0M9A5C4 A0A0K8VQ02 A0A034VCG9 A0A1A9WJC9 W8BEG9 A0A1B0F9U7 N6T871 A0A310SH56 A0A2W1BN22 A0A212F9W7 E0VJR3 A0A2S2NJR9 X1WTX5 A0A023EQ92 A0A0M3QYI5 B4QTH9 A0A232FBQ3
PDB
5YZV
E-value=0.0180329,
Score=90
Ontologies
KEGG
GO
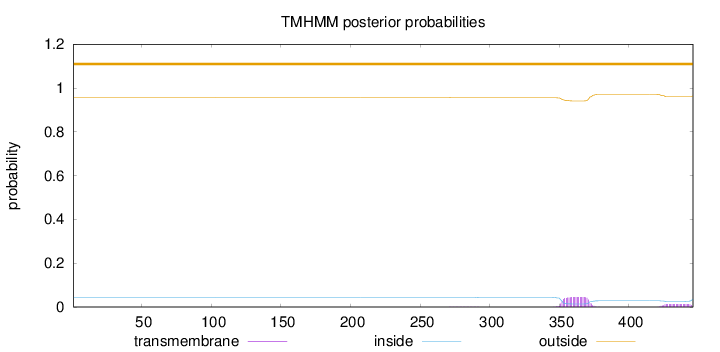
Topology
Length:
446
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.21246
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04359
outside
1 - 446
Population Genetic Test Statistics
Pi
17.6926
Theta
21.700414
Tajima's D
-0.71335
CLR
0.837149
CSRT
0.191090445477726
Interpretation
Uncertain
