Gene
KWMTBOMO12773
Pre Gene Modal
BGIBMGA007204
Annotation
PREDICTED:_uncharacterized_protein_LOC101740131_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.787
Sequence
CDS
ATGATTGCCAAAGAAACTAGTGATCACGAAAACTCTTCTAAATCCAATTTACAGAACGGTAATGAAATTAATGCAGATGGAGATTCAACGCATATAGATCACGATGAACATAAAGATGAAAAAAAAGTTGACACGTCCCATTACACAAAGAAAGAACCAATAGAATCGCCAGTCGAAAGAGTAAAGTTCGTGGGCATTGATGATGATGATGACGACGAAGACAGTTTTTATTCCGAGGAGTCATTCCAGAAAAAACTGCCTCCTATACCTGATGGGGGCTGGGGATGGGTGGTCGTTGCCTCTGCCTTCCTCGTATCCGCCTGTGTTGATGGTCTCGCTTTCTCGTTCGGACTCCTGCACGAAGAGTTCACAAGATATTTCGAAACGACACAATCTCAAACATCATTAATAGGAAGTCTATTTATAGCTACTCCACTGTTAGCCGGGCCAATAGCTAGCGCTATAGTAGACAGATTCGGGTGCAGAGTGATGACTATTATTGCCGGATTACTATCGACGATAGGATTTTTGTTAGCAGCGATTAGTAATTCAGTGTTGGTGTTATCTTTAACATTTGGTCTTCTCAGTGGCCTGGCCATGAGCATTCTCTACGTTACATCAGTAGTGGCTGTAGCGTTTTGGTTTGACAAAAGAAGGAACCTCGCTGTTTCACTGGCTTTGTGTGGAATGGGTTTCGGGACGGTGATTTATTCGCCGCTTACTCATTATTTTCTCCAAGTTTACGATTGGAGGAACACCGTAGTGCTACTTGCCGGTACATTATTGAATATGTGCGTTTGTGGGGCGCTGATGAGAAACCCTGAGTGGTTGGAAATTAAACAGAGAAGAGAGCGCAGGGTTGCTAAGGCGCGGTCGAAAAAGTCGTCAAGTGTCGCTTCATTGTCGTCGAGGTCTTTCGGAGAGGATTCGGAGTATCTGGGAGCGGAAGAACTGAAATCTCTGTTGCAGAGCGGCCAAAGTCCCGAATACATATTGGCGACGTTAGCGACAAGTATAGTCGAAGCTGAAAAATTGGAAGCCACGACTATGATGAACGCTGAACAAACATTCAAGAGAAATTATTCAGCGATACATTTGCCTACATTCATATTGCAGAACGAAATGGTCCCATCAGAAGTAATTGAAAAATTAATGGAAAACAAGAACCTATACAACATTGTCTTACAAAATTACCCAGATTCTATAACACGAAGGAAATCCGAGCTCAATGTTAATGTAGAAACTCCATCTACGAACATTGTTACAGAACCAGCGAAACAACACGTTGAGATAAAAGTTAAAAAACCTAAACATACACAAACAGAAAACGAGAAGCTTGAAGAAAAGAAAACTGTAGAAAGAAAGAAAAGTGTCGAAGTTAAAACAAAAATTCAGGGAACAGATGACAATGCCAAAAATAAAACTAATAAAGATGCGAAGTTACACGCGCAGCCTAGTTGGCTATGGCAAATCTCGACCGATCATCAGTACCTGCGAGACATGCCTTTGTATAGAAAAAAAATTATGCACCGCGGAGCCATGCTGAGTATTCCTAGATACAGACTGAGAGCGTCCTCTTTACCGGACATGTACAGGAACTCCATGTGGTCTTTTGAATCTGACTCGGATGATGAAATGGAATGGTACCACCGATTTTGGCAATCGACCAAAAACATGTTCGACTTCAGAATGCTGATGGAGCTACATTTCTTCTTGTTAAATTTGTCGTCAATGATACTGTCCGTGTGGTTCATCGTGCCGTACTTCTTCCTCAACTCTTACATGAAAGATTATAACGTCGAAGGCGGCGCTACTATGATCAGTATCATTGGAGTCGCCAGCAGCATTGGAATCGTAGCCCTCGGCTGGACTGGTGATCAACCGTGGGTCAACGTCACCAAAACCTACGCAATTTGCCTCATCGTCTGCGGTGTGTCCGTTGCCGCGTACCCCCTCTTCATCACCAACTATTGGATGCTATCCGTCATCTCCGCAGTCTTTGGGGTCACCTTCGCCAGCACGTATTCGTACACTCCAGCCATTCTAATGGAGCTGCTGCCGGTCGAGAACTTTACTGTGGGGTACGGCCTCATCCTGCTCAACGAAGGGATCGGTCACCTTATAGGACCACCGATCGGAGGGCTCCTCTACGACTTGACCGGAACCTGGGATCTGACGTTCTACATGGGCGGTCTGTGGATAGTCATCTCGGGGCTGTGCGTGGGGCTCGTGGACTTCACCAGAAACCGGCAGATGTGCGGAAGCGCTCCGCTACTGAAGGAGGCTGCCGCTTCGGACGGAGACGACAGCTCCGAAGAGTGA
Protein
MIAKETSDHENSSKSNLQNGNEINADGDSTHIDHDEHKDEKKVDTSHYTKKEPIESPVERVKFVGIDDDDDDEDSFYSEESFQKKLPPIPDGGWGWVVVASAFLVSACVDGLAFSFGLLHEEFTRYFETTQSQTSLIGSLFIATPLLAGPIASAIVDRFGCRVMTIIAGLLSTIGFLLAAISNSVLVLSLTFGLLSGLAMSILYVTSVVAVAFWFDKRRNLAVSLALCGMGFGTVIYSPLTHYFLQVYDWRNTVVLLAGTLLNMCVCGALMRNPEWLEIKQRRERRVAKARSKKSSSVASLSSRSFGEDSEYLGAEELKSLLQSGQSPEYILATLATSIVEAEKLEATTMMNAEQTFKRNYSAIHLPTFILQNEMVPSEVIEKLMENKNLYNIVLQNYPDSITRRKSELNVNVETPSTNIVTEPAKQHVEIKVKKPKHTQTENEKLEEKKTVERKKSVEVKTKIQGTDDNAKNKTNKDAKLHAQPSWLWQISTDHQYLRDMPLYRKKIMHRGAMLSIPRYRLRASSLPDMYRNSMWSFESDSDDEMEWYHRFWQSTKNMFDFRMLMELHFFLLNLSSMILSVWFIVPYFFLNSYMKDYNVEGGATMISIIGVASSIGIVALGWTGDQPWVNVTKTYAICLIVCGVSVAAYPLFITNYWMLSVISAVFGVTFASTYSYTPAILMELLPVENFTVGYGLILLNEGIGHLIGPPIGGLLYDLTGTWDLTFYMGGLWIVISGLCVGLVDFTRNRQMCGSAPLLKEAAASDGDDSSEE
Summary
Uniprot
H9JCF9
A0A2A4JUI4
A0A2W1BN40
A0A194QG55
A0A2H1VWU9
A0A0N1INZ7
+ More
A0A3S2NL29 H9JCA9 A0A0L7L314 A0A212FI86 A0A2A4IUF1 A0A2W1BRL4 A0A2H1V1L2 A0A3S2NAV0 H9J328 A0A194RIL6 A0A194PK99 A0A212EL73 A0A1B6MQ51 A0A1B6FAH8 A0A1B6CQY0 A0A1B6JKQ8 A0A1B6DDB5 A0A2J7QWR1 A0A0M9A4Z2 A0A2P8Y960 A0A1B6FEK1 E2B035 F4WBF5 A0A151K1W4 A0A2A3EMT5 A0A151X9S7 A0A087ZZF8 A0A1Y1K255 A0A151JP46 A0A232EY82 E0VFL1 A0A195B559 K7ISI1 E9JCL6 A0A310SCB8 D6WW59 A0A3L8DBS4 A0A026WKB9 J9JX09 A0A2S2NJJ7 A0A0N0P9U3 A0A0J7NFB9 N6U8D4 E2BIZ7 A0A154PIA3 N6U5M4 A0A194PIB8 A0A2H1VBZ1 A0A2W1C0X9 A0A195CUZ7 A0A437AX13 H9J327 A0A194RJQ5 A0A023EVZ8 A0A0L7KU96 A0A182H912 A0A154PI87 A0A023EVT0 E2B036 A0A1B6D3S6 A0A0J7KMG8 E2BIZ9 A0A0L7QNA7 A0A310S6P6 A0A212EL79 A0A087ZZF7 A0A224XKF4 A0A026WLG7 A0A195B5U1 A0A1Y1M0L3 T1HW00 A0A158NQA4 A0A1J1IHT6 A0A151XA28 A0A0A9XLI7 A0A1W4W469 A0A195CTG3 A0A224X9P2 A0A1W4W461 F4WBF6 A0A2S2QFW9 A0A232FAB7 A0A151JNR2 A0A1B6C2J0 A0A1B6GX81 E9JCL7 E0VFK5 U5ETA5 A0A2S2NID1 D6WW60 A0A1L8DE12 A0A1L8DEE9 A0A1L8DE24
A0A3S2NL29 H9JCA9 A0A0L7L314 A0A212FI86 A0A2A4IUF1 A0A2W1BRL4 A0A2H1V1L2 A0A3S2NAV0 H9J328 A0A194RIL6 A0A194PK99 A0A212EL73 A0A1B6MQ51 A0A1B6FAH8 A0A1B6CQY0 A0A1B6JKQ8 A0A1B6DDB5 A0A2J7QWR1 A0A0M9A4Z2 A0A2P8Y960 A0A1B6FEK1 E2B035 F4WBF5 A0A151K1W4 A0A2A3EMT5 A0A151X9S7 A0A087ZZF8 A0A1Y1K255 A0A151JP46 A0A232EY82 E0VFL1 A0A195B559 K7ISI1 E9JCL6 A0A310SCB8 D6WW59 A0A3L8DBS4 A0A026WKB9 J9JX09 A0A2S2NJJ7 A0A0N0P9U3 A0A0J7NFB9 N6U8D4 E2BIZ7 A0A154PIA3 N6U5M4 A0A194PIB8 A0A2H1VBZ1 A0A2W1C0X9 A0A195CUZ7 A0A437AX13 H9J327 A0A194RJQ5 A0A023EVZ8 A0A0L7KU96 A0A182H912 A0A154PI87 A0A023EVT0 E2B036 A0A1B6D3S6 A0A0J7KMG8 E2BIZ9 A0A0L7QNA7 A0A310S6P6 A0A212EL79 A0A087ZZF7 A0A224XKF4 A0A026WLG7 A0A195B5U1 A0A1Y1M0L3 T1HW00 A0A158NQA4 A0A1J1IHT6 A0A151XA28 A0A0A9XLI7 A0A1W4W469 A0A195CTG3 A0A224X9P2 A0A1W4W461 F4WBF6 A0A2S2QFW9 A0A232FAB7 A0A151JNR2 A0A1B6C2J0 A0A1B6GX81 E9JCL7 E0VFK5 U5ETA5 A0A2S2NID1 D6WW60 A0A1L8DE12 A0A1L8DEE9 A0A1L8DE24
Pubmed
EMBL
BABH01031610
NWSH01000554
PCG75685.1
KZ150013
PZC75044.1
KQ458981
+ More
KPJ04472.1 ODYU01004928 SOQ45305.1 KQ460882 KPJ11474.1 RSAL01000317 RVE42585.1 BABH01031609 JTDY01003280 KOB69810.1 AGBW02008415 OWR53453.1 NWSH01007033 PCG63126.1 KZ149921 PZC77732.1 ODYU01000260 SOQ34727.1 RSAL01000304 RVE42716.1 BABH01037649 KQ460124 KPJ17668.1 KQ459603 KPI93154.1 AGBW02014130 OWR42229.1 GEBQ01017028 GEBQ01016920 GEBQ01012615 GEBQ01001956 JAT22949.1 JAT23057.1 JAT27362.1 JAT38021.1 GECZ01022618 GECZ01008634 JAS47151.1 JAS61135.1 GEDC01021494 JAS15804.1 GECU01028486 GECU01022863 GECU01016348 GECU01008024 JAS79220.1 JAS84843.1 JAS91358.1 JAS99682.1 GEDC01013652 JAS23646.1 NEVH01009422 PNF33023.1 KQ435732 KOX77337.1 PYGN01000790 PSN40793.1 GECZ01021235 JAS48534.1 GL444372 EFN60961.1 GL888062 EGI68511.1 KQ981189 KYN45223.1 KZ288212 PBC32804.1 KQ982365 KYQ57127.1 GEZM01099141 JAV53466.1 KQ978820 KYN28133.1 NNAY01001665 OXU23277.1 DS235115 EEB12167.1 KQ976595 KYM79626.1 GL771849 EFZ09427.1 KQ779356 OAD51997.1 KQ971361 EFA08671.2 QOIP01000010 RLU17786.1 KK107167 EZA56497.1 ABLF02030750 GGMR01004731 MBY17350.1 KQ459435 KPJ01099.1 LBMM01005754 KMQ91230.1 APGK01038976 APGK01038977 APGK01038978 APGK01038979 KB740966 ENN76911.1 GL448543 EFN84342.1 KQ434924 KZC11585.1 APGK01038973 KB631578 ENN76910.1 ERL84404.1 KPI93156.1 ODYU01001747 SOQ38363.1 PZC77733.1 KQ977279 KYN03989.1 RVE42717.1 BABH01037651 KPJ17669.1 GAPW01000291 JAC13307.1 JTDY01005603 KOB66842.1 JXUM01119799 JXUM01119800 KQ566325 KXJ70232.1 KZC11586.1 GAPW01000290 JAC13308.1 EFN60962.1 GEDC01030362 GEDC01016970 JAS06936.1 JAS20328.1 LBMM01005457 KMQ91497.1 EFN84344.1 KQ414855 KOC60127.1 OAD51996.1 OWR42228.1 GFTR01007803 JAW08623.1 EZA56496.1 RLU17785.1 KYM79627.1 GEZM01042994 GEZM01042993 JAV79402.1 ACPB03014175 ADTU01022996 CVRI01000053 CRK99823.1 KYQ57128.1 GBHO01025619 GDHC01000641 JAG17985.1 JAQ17988.1 KYN03988.1 GFTR01007370 JAW09056.1 EGI68512.1 GGMS01007197 MBY76400.1 NNAY01000581 OXU27562.1 KYN28134.1 GEDC01029848 JAS07450.1 GECZ01002733 JAS67036.1 EFZ09435.1 EEB12161.1 GANO01004466 JAB55405.1 GGMR01004318 MBY16937.1 EFA08184.1 GFDF01009381 JAV04703.1 GFDF01009380 JAV04704.1 GFDF01009382 JAV04702.1
KPJ04472.1 ODYU01004928 SOQ45305.1 KQ460882 KPJ11474.1 RSAL01000317 RVE42585.1 BABH01031609 JTDY01003280 KOB69810.1 AGBW02008415 OWR53453.1 NWSH01007033 PCG63126.1 KZ149921 PZC77732.1 ODYU01000260 SOQ34727.1 RSAL01000304 RVE42716.1 BABH01037649 KQ460124 KPJ17668.1 KQ459603 KPI93154.1 AGBW02014130 OWR42229.1 GEBQ01017028 GEBQ01016920 GEBQ01012615 GEBQ01001956 JAT22949.1 JAT23057.1 JAT27362.1 JAT38021.1 GECZ01022618 GECZ01008634 JAS47151.1 JAS61135.1 GEDC01021494 JAS15804.1 GECU01028486 GECU01022863 GECU01016348 GECU01008024 JAS79220.1 JAS84843.1 JAS91358.1 JAS99682.1 GEDC01013652 JAS23646.1 NEVH01009422 PNF33023.1 KQ435732 KOX77337.1 PYGN01000790 PSN40793.1 GECZ01021235 JAS48534.1 GL444372 EFN60961.1 GL888062 EGI68511.1 KQ981189 KYN45223.1 KZ288212 PBC32804.1 KQ982365 KYQ57127.1 GEZM01099141 JAV53466.1 KQ978820 KYN28133.1 NNAY01001665 OXU23277.1 DS235115 EEB12167.1 KQ976595 KYM79626.1 GL771849 EFZ09427.1 KQ779356 OAD51997.1 KQ971361 EFA08671.2 QOIP01000010 RLU17786.1 KK107167 EZA56497.1 ABLF02030750 GGMR01004731 MBY17350.1 KQ459435 KPJ01099.1 LBMM01005754 KMQ91230.1 APGK01038976 APGK01038977 APGK01038978 APGK01038979 KB740966 ENN76911.1 GL448543 EFN84342.1 KQ434924 KZC11585.1 APGK01038973 KB631578 ENN76910.1 ERL84404.1 KPI93156.1 ODYU01001747 SOQ38363.1 PZC77733.1 KQ977279 KYN03989.1 RVE42717.1 BABH01037651 KPJ17669.1 GAPW01000291 JAC13307.1 JTDY01005603 KOB66842.1 JXUM01119799 JXUM01119800 KQ566325 KXJ70232.1 KZC11586.1 GAPW01000290 JAC13308.1 EFN60962.1 GEDC01030362 GEDC01016970 JAS06936.1 JAS20328.1 LBMM01005457 KMQ91497.1 EFN84344.1 KQ414855 KOC60127.1 OAD51996.1 OWR42228.1 GFTR01007803 JAW08623.1 EZA56496.1 RLU17785.1 KYM79627.1 GEZM01042994 GEZM01042993 JAV79402.1 ACPB03014175 ADTU01022996 CVRI01000053 CRK99823.1 KYQ57128.1 GBHO01025619 GDHC01000641 JAG17985.1 JAQ17988.1 KYN03988.1 GFTR01007370 JAW09056.1 EGI68512.1 GGMS01007197 MBY76400.1 NNAY01000581 OXU27562.1 KYN28134.1 GEDC01029848 JAS07450.1 GECZ01002733 JAS67036.1 EFZ09435.1 EEB12161.1 GANO01004466 JAB55405.1 GGMR01004318 MBY16937.1 EFA08184.1 GFDF01009381 JAV04703.1 GFDF01009380 JAV04704.1 GFDF01009382 JAV04702.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000037510
+ More
UP000007151 UP000235965 UP000053105 UP000245037 UP000000311 UP000007755 UP000078541 UP000242457 UP000075809 UP000005203 UP000078492 UP000215335 UP000009046 UP000078540 UP000002358 UP000007266 UP000279307 UP000053097 UP000007819 UP000036403 UP000019118 UP000008237 UP000076502 UP000030742 UP000078542 UP000069940 UP000249989 UP000053825 UP000015103 UP000005205 UP000183832 UP000192223
UP000007151 UP000235965 UP000053105 UP000245037 UP000000311 UP000007755 UP000078541 UP000242457 UP000075809 UP000005203 UP000078492 UP000215335 UP000009046 UP000078540 UP000002358 UP000007266 UP000279307 UP000053097 UP000007819 UP000036403 UP000019118 UP000008237 UP000076502 UP000030742 UP000078542 UP000069940 UP000249989 UP000053825 UP000015103 UP000005205 UP000183832 UP000192223
Pfam
PF07690 MFS_1
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JCF9
A0A2A4JUI4
A0A2W1BN40
A0A194QG55
A0A2H1VWU9
A0A0N1INZ7
+ More
A0A3S2NL29 H9JCA9 A0A0L7L314 A0A212FI86 A0A2A4IUF1 A0A2W1BRL4 A0A2H1V1L2 A0A3S2NAV0 H9J328 A0A194RIL6 A0A194PK99 A0A212EL73 A0A1B6MQ51 A0A1B6FAH8 A0A1B6CQY0 A0A1B6JKQ8 A0A1B6DDB5 A0A2J7QWR1 A0A0M9A4Z2 A0A2P8Y960 A0A1B6FEK1 E2B035 F4WBF5 A0A151K1W4 A0A2A3EMT5 A0A151X9S7 A0A087ZZF8 A0A1Y1K255 A0A151JP46 A0A232EY82 E0VFL1 A0A195B559 K7ISI1 E9JCL6 A0A310SCB8 D6WW59 A0A3L8DBS4 A0A026WKB9 J9JX09 A0A2S2NJJ7 A0A0N0P9U3 A0A0J7NFB9 N6U8D4 E2BIZ7 A0A154PIA3 N6U5M4 A0A194PIB8 A0A2H1VBZ1 A0A2W1C0X9 A0A195CUZ7 A0A437AX13 H9J327 A0A194RJQ5 A0A023EVZ8 A0A0L7KU96 A0A182H912 A0A154PI87 A0A023EVT0 E2B036 A0A1B6D3S6 A0A0J7KMG8 E2BIZ9 A0A0L7QNA7 A0A310S6P6 A0A212EL79 A0A087ZZF7 A0A224XKF4 A0A026WLG7 A0A195B5U1 A0A1Y1M0L3 T1HW00 A0A158NQA4 A0A1J1IHT6 A0A151XA28 A0A0A9XLI7 A0A1W4W469 A0A195CTG3 A0A224X9P2 A0A1W4W461 F4WBF6 A0A2S2QFW9 A0A232FAB7 A0A151JNR2 A0A1B6C2J0 A0A1B6GX81 E9JCL7 E0VFK5 U5ETA5 A0A2S2NID1 D6WW60 A0A1L8DE12 A0A1L8DEE9 A0A1L8DE24
A0A3S2NL29 H9JCA9 A0A0L7L314 A0A212FI86 A0A2A4IUF1 A0A2W1BRL4 A0A2H1V1L2 A0A3S2NAV0 H9J328 A0A194RIL6 A0A194PK99 A0A212EL73 A0A1B6MQ51 A0A1B6FAH8 A0A1B6CQY0 A0A1B6JKQ8 A0A1B6DDB5 A0A2J7QWR1 A0A0M9A4Z2 A0A2P8Y960 A0A1B6FEK1 E2B035 F4WBF5 A0A151K1W4 A0A2A3EMT5 A0A151X9S7 A0A087ZZF8 A0A1Y1K255 A0A151JP46 A0A232EY82 E0VFL1 A0A195B559 K7ISI1 E9JCL6 A0A310SCB8 D6WW59 A0A3L8DBS4 A0A026WKB9 J9JX09 A0A2S2NJJ7 A0A0N0P9U3 A0A0J7NFB9 N6U8D4 E2BIZ7 A0A154PIA3 N6U5M4 A0A194PIB8 A0A2H1VBZ1 A0A2W1C0X9 A0A195CUZ7 A0A437AX13 H9J327 A0A194RJQ5 A0A023EVZ8 A0A0L7KU96 A0A182H912 A0A154PI87 A0A023EVT0 E2B036 A0A1B6D3S6 A0A0J7KMG8 E2BIZ9 A0A0L7QNA7 A0A310S6P6 A0A212EL79 A0A087ZZF7 A0A224XKF4 A0A026WLG7 A0A195B5U1 A0A1Y1M0L3 T1HW00 A0A158NQA4 A0A1J1IHT6 A0A151XA28 A0A0A9XLI7 A0A1W4W469 A0A195CTG3 A0A224X9P2 A0A1W4W461 F4WBF6 A0A2S2QFW9 A0A232FAB7 A0A151JNR2 A0A1B6C2J0 A0A1B6GX81 E9JCL7 E0VFK5 U5ETA5 A0A2S2NID1 D6WW60 A0A1L8DE12 A0A1L8DEE9 A0A1L8DE24
Ontologies
GO
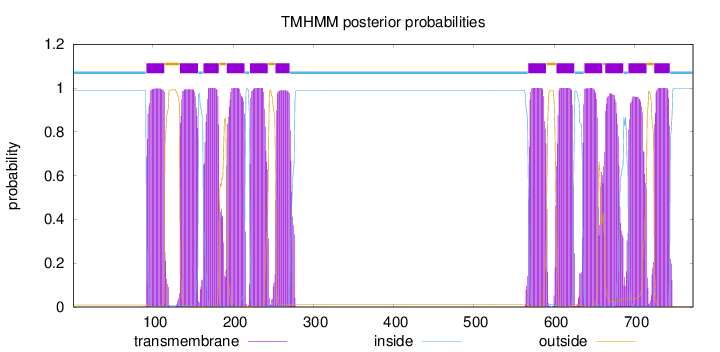
Topology
Length:
773
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
260.38384
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99039
inside
1 - 91
TMhelix
92 - 114
outside
115 - 133
TMhelix
134 - 156
inside
157 - 162
TMhelix
163 - 182
outside
183 - 191
TMhelix
192 - 214
inside
215 - 220
TMhelix
221 - 243
outside
244 - 252
TMhelix
253 - 270
inside
271 - 567
TMhelix
568 - 590
outside
591 - 602
TMhelix
603 - 625
inside
626 - 637
TMhelix
638 - 660
outside
661 - 663
TMhelix
664 - 686
inside
687 - 692
TMhelix
693 - 715
outside
716 - 724
TMhelix
725 - 744
inside
745 - 773
Population Genetic Test Statistics
Pi
19.946194
Theta
21.144134
Tajima's D
-0.491413
CLR
10.721381
CSRT
0.247387630618469
Interpretation
Uncertain
