Gene
KWMTBOMO12764
Pre Gene Modal
BGIBMGA007158
Annotation
PREDICTED:_phosphatidylinositol_3-kinase_catalytic_subunit_type_3_[Bombyx_mori]
Full name
Phosphatidylinositol 3-kinase catalytic subunit type 3
Alternative Name
Phosphoinositide-3-kinase class 3
Location in the cell
Cytoplasmic Reliability : 1.217 Mitochondrial Reliability : 1.399 Nuclear Reliability : 1.113
Sequence
CDS
ATGAACGAAATAGACGACAAGTTTTATTATGTTTACAGTTCATCTTTGGATTACAAGGTCGAAATAAAAATTGGTACCCTTGAAGGCAAGAGAACTAAACCGGAATATGACAAACTACTATCAGATCCGATGTTGAAATTCTCAGGACTTTACCAAGATGGATGCTCAGACTTGTACGTCACCTGCCAAATCCTGAGTGATGGAGAACCTCTGGCGCTACCCGTCATGACCACTTATAAGGCATTCACTACTCGATGGAATTGGAATGAATGGGTAACCCTTCCGGTGTACTACAGTGATTTGCCCCGTAATGCTATACTAGCTATGACTATCTACGACTGTGCCGGTCCCGGTAAAGTGATGGTGGTCGGTGGCACGACCATATCGATATTTGGGAAGCACGGCATGCTGAGACAGGGGATGTTCGATCTGCGAGTGTGGCCCGGGGTTGAGGGCAGCGAGAAGACTCCGGGGAAGGCTCCGGACCGCGGCAAGGAGCAGATGCAGCGACTTGCGAAGCTCGCCAAGAAACACAGAAACGGACATTTGCCCAAGGTAGACTGGCTGGACAGGCTGACGTTTCGTGAAATCGAGATGATAAATGAAAAGGAGAAGCGAAGTTCGAATTATATGTATCTGATGGTGGAATTCCCGAGTGTTCAGATCGACGGCATCAATCACGCAGTTGTGTACTACGAACAGGACGGCGATGAGATGTACCAGTTCAGAGCTCAGGCCGAGCTGGTCACCGTACCGGATTGCGAAATATTGAAGGAGAACATAGTGGAGAGCAAGCAGCACCGGCTGGCGCGGTCGCTGCGCGCGGGGCAGTCGGCGCGCGACGCCAAGCCCTCCGCGCTCGAGCGGGACCAGCTCGCCGCGCTGCTGGCCGCGCCGCCCGCCCGCGCCGCGCCCGCCCTGCACCAGGACCTGCTGTGGAAGTTCCGGTTTTACTTGTCGTCGCACCGCCGCGCCCTCACCAAGTTCCTGGAATGCGTCAATTGGAATAGACCTGGCGAAGTGCGTCAGGCCCTCGCCATGATGAGGCAGTGGGCGCCCATGGACGTGGAAGATGCGTTAGAGCTGCTGACTCCGAAGTTCACGCATCCCGCTGTTAGGAAATACGCGATATCGAGATTGAGACAGGCTCCCGACGAGGATCTGCTGCTGTACCTGCTGCAACTGGTCCAGGCCCTGAAGTACGAGAGCTACTCGGAAATACACGAGGAGTACGCGCGCACGTGCGGCAAGGACATGCCGCTACCCGTGGAGCTGGACCCTCGCCTGGGGTACAGCCACCGAGACACTCTGCTGGCCACCTCGGCTCTGAGCGACGTGTCCGCGACCAGCCGGGACCCGGACGGCGCGGGGGGACAGGGGGGACCGGGGGGACCCACCGGGGGACAGACCCCGGGGACCGCCGCGCCCAGCGATGACGTCACGTCTGGTGGCGAGGAGCAGGGCAGCCTGGCGTGGTTCCTGATCGACCGCGCGTGCCGCAACCCGCAGCTGGCCAACTACCTGTACTGGTACCTGCTGGTGGAGTGCGAGGACGCGCCCGAGCCGCCGCCCCCGCGACACATGTACCTCGCCGTCATGCGCACCTTCAGCAGACGACTCGCCGACGGTAGCAGCTCGCACCAGCGCACGCGCTCGTTCCTGGCGCGGCAGCAGGTGTTCATCGACAAGCTGGTGCGGCTGGTGCGCTCCGTGGCGCGGGAGAGCGGCAGCCGCGCGCGCAAGGCCCAGCGCCTGCAGCAGCTGCTCGCAGACCCCGACGCCAACAAGTTCAACTTCGCCAGCTTCGAGCCCATGCCCTTCCCGCTCGACCCCAGCGTCACCATCAAAGGGATCGTCGCCAAGAAGGCCAGCCTCTTCAAGTCCGCCCTCATGCCCACCAAGCTCACCTTCCTCACGACCGAGGGCGTCGAGTACGATGCCATCTTCAAGTACGGCGACGACCTGCGCCAAGACCAGCTGATCCTGCAGATGATCACGCTGATGGACAAGCTGCTGAGGCGCGAGAACCTGGACCTGAAGCTGACGCCGTACCGCGTGCTGGCCACCAGCTCCAAGCACGGCTTCCTGCAGTTCGTGGAGTCCGTGACGGTGGCGGAGGCGCTGGCCACGGAGGGCAGCATACAGAACTTCTTCAGGAAGCACAACCCGTGCGAGGGCGCGCCCTACGGCATCAAGCCCGAGACCATGGACACGTACATCCGCAGCTGCGCCGGATACTGCGTCATCACCTATCTGCTCGGTGTGGGTGACCGGCATCTGGACAACCTGCTGCTGAGCCGCAGCGGCGCGCTGTTCCACATCGACTTCGGCTACATCCTGGGTCGGGATCCGAAGCCGCTTCCGCCACCCATGAAGCTCAGCAAGGAGATGGTGGAGGCCATGGGCGGGGTCAACTCGGACCACTACCACGCCTTCCGCAAGCTCACCTACACCGCCTTCCTGCATCTCAGACGTCATGCAAACCTGATGCTGAACCTGTTCTCGCTGATGGTGGACGCCTCCGTCCCGGACATTGCGCTGGAGCCGGACAAGGCCGTCAAGAAAGTGCAGGATAAGTTCAGGCTGGATCTCGGCGACGAGGAAGCCATTCACTACCTGCAGAATCTCTTGGACATGTCAGTGACAGCTGTCATGGCGGTCCTCGTCGAACAATTCCACAAGTTCGCTCAATACTGGAGGAAATGA
Protein
MNEIDDKFYYVYSSSLDYKVEIKIGTLEGKRTKPEYDKLLSDPMLKFSGLYQDGCSDLYVTCQILSDGEPLALPVMTTYKAFTTRWNWNEWVTLPVYYSDLPRNAILAMTIYDCAGPGKVMVVGGTTISIFGKHGMLRQGMFDLRVWPGVEGSEKTPGKAPDRGKEQMQRLAKLAKKHRNGHLPKVDWLDRLTFREIEMINEKEKRSSNYMYLMVEFPSVQIDGINHAVVYYEQDGDEMYQFRAQAELVTVPDCEILKENIVESKQHRLARSLRAGQSARDAKPSALERDQLAALLAAPPARAAPALHQDLLWKFRFYLSSHRRALTKFLECVNWNRPGEVRQALAMMRQWAPMDVEDALELLTPKFTHPAVRKYAISRLRQAPDEDLLLYLLQLVQALKYESYSEIHEEYARTCGKDMPLPVELDPRLGYSHRDTLLATSALSDVSATSRDPDGAGGQGGPGGPTGGQTPGTAAPSDDVTSGGEEQGSLAWFLIDRACRNPQLANYLYWYLLVECEDAPEPPPPRHMYLAVMRTFSRRLADGSSSHQRTRSFLARQQVFIDKLVRLVRSVARESGSRARKAQRLQQLLADPDANKFNFASFEPMPFPLDPSVTIKGIVAKKASLFKSALMPTKLTFLTTEGVEYDAIFKYGDDLRQDQLILQMITLMDKLLRRENLDLKLTPYRVLATSSKHGFLQFVESVTVAEALATEGSIQNFFRKHNPCEGAPYGIKPETMDTYIRSCAGYCVITYLLGVGDRHLDNLLLSRSGALFHIDFGYILGRDPKPLPPPMKLSKEMVEAMGGVNSDHYHAFRKLTYTAFLHLRRHANLMLNLFSLMVDASVPDIALEPDKAVKKVQDKFRLDLGDEEAIHYLQNLLDMSVTAVMAVLVEQFHKFAQYWRK
Summary
Description
Catalytic subunit of the PI3K complex that mediates formation of phosphatidylinositol 3-phosphate; different complex forms are believed to play a role in multiple membrane trafficking pathways. Involved in the transport of lysosomal enzyme precursors to lysosomes. Required for transport from early to late endosomes (By similarity).
Catalytic subunit of the PI3K complex that mediates formation of phosphatidylinositol 3-phosphate; different complex forms are believed to play a role in multiple membrane trafficking pathways: PI3KC3-C1 is involved in initiation of autophagosomes and PI3KC3-C2 in maturation of autophagosomes and endocytosis. Involved in regulation of degradative endocytic trafficking and required for the abcission step in cytokinesis, probably in the context of PI3KC3-C2. Involved in the transport of lysosomal enzyme precursors to lysosomes. Required for transport from early to late endosomes (By similarity).
Catalytic subunit of the PI3K complex that mediates formation of phosphatidylinositol 3-phosphate; different complex forms are believed to play a role in multiple membrane trafficking pathways: PI3KC3-C1 is involved in initiation of autophagosomes and PI3KC3-C2 in maturation of autophagosomes and endocytosis. Involved in regulation of degradative endocytic trafficking and required for the abcission step in cytokinesis, probably in the context of PI3KC3-C2. Involved in the transport of lysosomal enzyme precursors to lysosomes. Required for transport from early to late endosomes (By similarity).
Catalytic Activity
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol) + ATP = a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol 3-phosphate) + ADP + H(+)
Cofactor
Mn(2+)
Subunit
Component of the PI3K (PI3KC3/PI3K-III/class III phosphatidylinositol 3-kinase) complex the core of which is composed of the catalytic subunit pik3c3, the regulatory subunit pik3r4 and becn1 associating with additional regulatory/auxilliary subunits to form alternative complex forms.
Component of the PI3K (PI3KC3/PI3K-III/class III phosphatidylinositol 3-kinase) complex the core of which is composed of the catalytic subunit PIK3C3, the regulatory subunit PIK3R4 and BECN1 associating with additional regulatory/auxilliary subunits to form alternative complex forms. Alternative complex forms containing a forth regulatory subunit in a mutually exclusive manner are: the PI3K complex I (PI3KC3-C1) containing ATG14, and the PI3K complex II (PI3KC3-C2) containing UVRAG. PI3KC3-C1 displays a V-shaped architecture with PIK3R4 serving as a bridge between PIK3C3 and the ATG14:BECN1 subcomplex (By similarity). Both, PI3KC3-C1 and PI3KC3-C2, can associate with further regulatory subunits such as RUBCN, SH3GLB1/Bif-1 and AMBRA1 (PubMed:17589504). PI3KC3-C1 probably associates with PIK3CB (PubMed:21059846). Interacts with RAB7A in the presence of PIK3R4 (By similarity). Interacts with AMBRA1 (PubMed:17589504). Interacts with BECN1P1/BECN2 (By similarity). Interacts with SLAMF1 (PubMed:22493499). May be a component of a complex composed of RAB5A (in GDP-bound form), DYN2 and PIK3C3 (By similarity). Interacts with NCKAP1L (By similarity).
Component of the PI3K (PI3KC3/PI3K-III/class III phosphatidylinositol 3-kinase) complex the core of which is composed of the catalytic subunit PIK3C3, the regulatory subunit PIK3R4 and BECN1 associating with additional regulatory/auxilliary subunits to form alternative complex forms. Alternative complex forms containing a forth regulatory subunit in a mutually exclusive manner are: the PI3K complex I (PI3KC3-C1) containing ATG14, and the PI3K complex II (PI3KC3-C2) containing UVRAG. PI3KC3-C1 displays a V-shaped architecture with PIK3R4 serving as a bridge between PIK3C3 and the ATG14:BECN1 subcomplex (By similarity). Both, PI3KC3-C1 and PI3KC3-C2, can associate with further regulatory subunits such as RUBCN, SH3GLB1/Bif-1 and AMBRA1 (PubMed:17589504). PI3KC3-C1 probably associates with PIK3CB (PubMed:21059846). Interacts with RAB7A in the presence of PIK3R4 (By similarity). Interacts with AMBRA1 (PubMed:17589504). Interacts with BECN1P1/BECN2 (By similarity). Interacts with SLAMF1 (PubMed:22493499). May be a component of a complex composed of RAB5A (in GDP-bound form), DYN2 and PIK3C3 (By similarity). Interacts with NCKAP1L (By similarity).
Similarity
Belongs to the PI3/PI4-kinase family.
Keywords
ATP-binding
Autophagy
Cell cycle
Cell division
Kinase
Manganese
Nucleotide-binding
Transferase
Alternative splicing
Complete proteome
Cytoplasmic vesicle
Endosome
Phosphoprotein
Reference proteome
Feature
chain Phosphatidylinositol 3-kinase catalytic subunit type 3
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A194QLY7
A0A212F9V4
H9JCB3
A0A2H1VUC4
A0A1Y1KF83
A0A2J7PWP0
+ More
A0A067RKC6 A0A0P4VM95 T1HML4 A0A023F599 D6W9A3 A0A195BWH8 A0A154PFT4 E2B101 A0A195D9L4 A0A0L7RF14 A0A151WST9 A0A195CQ30 A0A195FQL7 A0A2R7VZ13 A0A3L8D828 A0A2A3EGH2 E0VQ65 E9IDA9 A0A2J7PWM9 A0A1S4EPX6 A0A2S2Q897 T1JEU2 J9K3U7 A0A310SPJ0 A0A2H8TFV5 A0A2P8XVN6 A0A1L8DMC6 B0W5Z6 C3Y1V6 A0A1Q3FDS2 A0A146P539 A0A3P8UT11 A0A3Q2FWL0 A0A2U9B189 A0A1L8I323 Q6AZN6 A0A3B4WVD8 A0A3B4TW17 G3Q5W9 A0A1A9WPA1 A0A3Q2PAN6 A0A3Q1FZN2 A0A3P8S0F3 A0A1W4ZW98 H2LUA0 A0A3P9LW04 A0A1B0CTU6 A0A1W4ZNA0 A0A1U8CKK8 A0A2I4CLW9 A0A3B3C9C2 A0A3Q2E6U0 A0A3Q3AKW4 G3Q5X0 Q6PF93 A0A3B5Q9A0 A0A1V4KTV3 I3K560 A0A3B3WQT0 A0A1U7RYK9 A0A3B3W1B4 A0A3S2PT87 A0A1J1ITM6 A0A147ASV9 A0A1A8RSW8 G1KPF8 F1Q9F3 A0A1A8NAL0 A0A1A8I9L5 A0A1A8UHN0 G1SX90 A0A3P9IUA7 A0A3Q3KSY3 A0A1A8C650 A0A146Y7U9 A0A3B4X232 A0A151MWA8 A0A3B1J562 A0A3Q2UEL4 A0A1S3SDV9 A0A3B4EA32 T1E4S4 A0A402EVR1 A0A1A8H9A0 A0A0F8B087 A0A060WR34 A0A3B3HSF3 A0A3P9KBE9 A0A1S3SDV8 J3SCI6 A0A3B4F817 A0A2I4CLX1 A0A3Q2VFU5 K9IP63
A0A067RKC6 A0A0P4VM95 T1HML4 A0A023F599 D6W9A3 A0A195BWH8 A0A154PFT4 E2B101 A0A195D9L4 A0A0L7RF14 A0A151WST9 A0A195CQ30 A0A195FQL7 A0A2R7VZ13 A0A3L8D828 A0A2A3EGH2 E0VQ65 E9IDA9 A0A2J7PWM9 A0A1S4EPX6 A0A2S2Q897 T1JEU2 J9K3U7 A0A310SPJ0 A0A2H8TFV5 A0A2P8XVN6 A0A1L8DMC6 B0W5Z6 C3Y1V6 A0A1Q3FDS2 A0A146P539 A0A3P8UT11 A0A3Q2FWL0 A0A2U9B189 A0A1L8I323 Q6AZN6 A0A3B4WVD8 A0A3B4TW17 G3Q5W9 A0A1A9WPA1 A0A3Q2PAN6 A0A3Q1FZN2 A0A3P8S0F3 A0A1W4ZW98 H2LUA0 A0A3P9LW04 A0A1B0CTU6 A0A1W4ZNA0 A0A1U8CKK8 A0A2I4CLW9 A0A3B3C9C2 A0A3Q2E6U0 A0A3Q3AKW4 G3Q5X0 Q6PF93 A0A3B5Q9A0 A0A1V4KTV3 I3K560 A0A3B3WQT0 A0A1U7RYK9 A0A3B3W1B4 A0A3S2PT87 A0A1J1ITM6 A0A147ASV9 A0A1A8RSW8 G1KPF8 F1Q9F3 A0A1A8NAL0 A0A1A8I9L5 A0A1A8UHN0 G1SX90 A0A3P9IUA7 A0A3Q3KSY3 A0A1A8C650 A0A146Y7U9 A0A3B4X232 A0A151MWA8 A0A3B1J562 A0A3Q2UEL4 A0A1S3SDV9 A0A3B4EA32 T1E4S4 A0A402EVR1 A0A1A8H9A0 A0A0F8B087 A0A060WR34 A0A3B3HSF3 A0A3P9KBE9 A0A1S3SDV8 J3SCI6 A0A3B4F817 A0A2I4CLX1 A0A3Q2VFU5 K9IP63
EC Number
2.7.1.137
Pubmed
26354079
22118469
19121390
28004739
24845553
27129103
+ More
25474469 18362917 19820115 20798317 30249741 20566863 21282665 29403074 18563158 24487278 27762356 17554307 29451363 15489334 17589504 21183079 21059846 22493499 23332761 24089209 23542700 25186727 23594743 21993624 22293439 25329095 15592404 23758969 25835551 24755649 23025625
25474469 18362917 19820115 20798317 30249741 20566863 21282665 29403074 18563158 24487278 27762356 17554307 29451363 15489334 17589504 21183079 21059846 22493499 23332761 24089209 23542700 25186727 23594743 21993624 22293439 25329095 15592404 23758969 25835551 24755649 23025625
EMBL
KQ458981
KPJ04466.1
AGBW02009556
OWR50514.1
BABH01031593
BABH01031594
+ More
ODYU01004213 SOQ43844.1 GEZM01087202 JAV58850.1 NEVH01020871 PNF20745.1 KK852460 KDR23478.1 GDKW01000953 JAI55642.1 ACPB03020499 GBBI01002434 JAC16278.1 KQ971312 EEZ98191.1 KQ976401 KYM92635.1 KQ434886 KZC10178.1 GL444767 EFN60627.1 KQ981082 KYN09610.1 KQ414606 KOC69577.1 KQ982769 KYQ50916.1 KQ977444 KYN02740.1 KQ981335 KYN42592.1 KK854182 PTY12724.1 QOIP01000011 RLU16647.1 KZ288267 PBC30101.1 DS235389 EEB15521.1 GL762454 EFZ21498.1 PNF20743.1 GGMS01004752 MBY73955.1 JH432130 ABLF02026968 KQ762031 OAD56362.1 GFXV01001144 MBW12949.1 PYGN01001278 PSN36071.1 GFDF01006570 JAV07514.1 DS231845 EDS35983.1 GG666480 EEN65846.1 GFDL01009334 JAV25711.1 GCES01147447 JAQ38875.1 CP026244 AWO97541.1 CM004466 OCU02735.1 BC077528 AJWK01027997 BC024675 BC057678 LSYS01001587 OPJ87851.1 AERX01017946 CM012454 RVE60403.1 CVRI01000057 CRL02454.1 GCES01004660 JAR81663.1 HAEH01003759 HAEI01009684 SBS08463.1 BX510349 HAEF01021582 HAEG01001868 SBR66118.1 HAED01007585 HAEE01003542 SBQ93797.1 HADY01004517 HAEJ01007133 SBS47590.1 AAGW02032208 AAGW02032209 AAGW02032210 HADZ01011436 HAEA01007987 SBP75377.1 GCES01036072 JAR50251.1 AKHW03004724 KYO28843.1 AC189108 AC192000 GAAZ01001775 JAA96168.1 BDOT01000049 GCF48210.1 HAEB01002391 HAEC01011695 SBQ79912.1 KQ042129 KKF18716.1 FR904686 CDQ69823.1 JU174894 GBEX01002474 AFJ50420.1 JAI12086.1 GABZ01004243 JAA49282.1
ODYU01004213 SOQ43844.1 GEZM01087202 JAV58850.1 NEVH01020871 PNF20745.1 KK852460 KDR23478.1 GDKW01000953 JAI55642.1 ACPB03020499 GBBI01002434 JAC16278.1 KQ971312 EEZ98191.1 KQ976401 KYM92635.1 KQ434886 KZC10178.1 GL444767 EFN60627.1 KQ981082 KYN09610.1 KQ414606 KOC69577.1 KQ982769 KYQ50916.1 KQ977444 KYN02740.1 KQ981335 KYN42592.1 KK854182 PTY12724.1 QOIP01000011 RLU16647.1 KZ288267 PBC30101.1 DS235389 EEB15521.1 GL762454 EFZ21498.1 PNF20743.1 GGMS01004752 MBY73955.1 JH432130 ABLF02026968 KQ762031 OAD56362.1 GFXV01001144 MBW12949.1 PYGN01001278 PSN36071.1 GFDF01006570 JAV07514.1 DS231845 EDS35983.1 GG666480 EEN65846.1 GFDL01009334 JAV25711.1 GCES01147447 JAQ38875.1 CP026244 AWO97541.1 CM004466 OCU02735.1 BC077528 AJWK01027997 BC024675 BC057678 LSYS01001587 OPJ87851.1 AERX01017946 CM012454 RVE60403.1 CVRI01000057 CRL02454.1 GCES01004660 JAR81663.1 HAEH01003759 HAEI01009684 SBS08463.1 BX510349 HAEF01021582 HAEG01001868 SBR66118.1 HAED01007585 HAEE01003542 SBQ93797.1 HADY01004517 HAEJ01007133 SBS47590.1 AAGW02032208 AAGW02032209 AAGW02032210 HADZ01011436 HAEA01007987 SBP75377.1 GCES01036072 JAR50251.1 AKHW03004724 KYO28843.1 AC189108 AC192000 GAAZ01001775 JAA96168.1 BDOT01000049 GCF48210.1 HAEB01002391 HAEC01011695 SBQ79912.1 KQ042129 KKF18716.1 FR904686 CDQ69823.1 JU174894 GBEX01002474 AFJ50420.1 JAI12086.1 GABZ01004243 JAA49282.1
Proteomes
UP000053268
UP000007151
UP000005204
UP000235965
UP000027135
UP000015103
+ More
UP000007266 UP000078540 UP000076502 UP000000311 UP000078492 UP000053825 UP000075809 UP000078542 UP000078541 UP000279307 UP000242457 UP000009046 UP000079169 UP000007819 UP000245037 UP000002320 UP000001554 UP000265000 UP000265120 UP000265020 UP000246464 UP000186698 UP000261360 UP000261420 UP000007635 UP000091820 UP000257200 UP000265080 UP000192224 UP000001038 UP000265180 UP000092461 UP000189706 UP000192220 UP000261560 UP000264800 UP000000589 UP000002852 UP000190648 UP000005207 UP000261480 UP000189705 UP000261500 UP000183832 UP000001646 UP000000437 UP000001811 UP000265200 UP000261660 UP000050525 UP000018467 UP000000539 UP000087266 UP000261440 UP000288954 UP000193380 UP000261460 UP000264840
UP000007266 UP000078540 UP000076502 UP000000311 UP000078492 UP000053825 UP000075809 UP000078542 UP000078541 UP000279307 UP000242457 UP000009046 UP000079169 UP000007819 UP000245037 UP000002320 UP000001554 UP000265000 UP000265120 UP000265020 UP000246464 UP000186698 UP000261360 UP000261420 UP000007635 UP000091820 UP000257200 UP000265080 UP000192224 UP000001038 UP000265180 UP000092461 UP000189706 UP000192220 UP000261560 UP000264800 UP000000589 UP000002852 UP000190648 UP000005207 UP000261480 UP000189705 UP000261500 UP000183832 UP000001646 UP000000437 UP000001811 UP000265200 UP000261660 UP000050525 UP000018467 UP000000539 UP000087266 UP000261440 UP000288954 UP000193380 UP000261460 UP000264840
Pfam
Interpro
IPR008290
PI3K_Vps34
+ More
IPR036940 PI3/4_kinase_cat_sf
IPR015433 PI_Kinase
IPR016024 ARM-type_fold
IPR035892 C2_domain_sf
IPR001263 PI3K_accessory_dom
IPR042236 PI3K_accessory_sf
IPR002420 PI3K_C2_dom
IPR011009 Kinase-like_dom_sf
IPR000403 PI3/4_kinase_cat_dom
IPR018936 PI3/4_kinase_CS
IPR001098 DNA-dir_DNA_pol_A_palm_dom
IPR002298 DNA_polymerase_A
IPR036322 WD40_repeat_dom_sf
IPR007074 LicD_fam
IPR015399 DUF1977_DnaJ-like
IPR022587 MTMR12-like_C
IPR029021 Prot-tyrosine_phosphatase-like
IPR036869 J_dom_sf
IPR010569 Myotubularin-like_Pase_dom
IPR001623 DnaJ_domain
IPR036940 PI3/4_kinase_cat_sf
IPR015433 PI_Kinase
IPR016024 ARM-type_fold
IPR035892 C2_domain_sf
IPR001263 PI3K_accessory_dom
IPR042236 PI3K_accessory_sf
IPR002420 PI3K_C2_dom
IPR011009 Kinase-like_dom_sf
IPR000403 PI3/4_kinase_cat_dom
IPR018936 PI3/4_kinase_CS
IPR001098 DNA-dir_DNA_pol_A_palm_dom
IPR002298 DNA_polymerase_A
IPR036322 WD40_repeat_dom_sf
IPR007074 LicD_fam
IPR015399 DUF1977_DnaJ-like
IPR022587 MTMR12-like_C
IPR029021 Prot-tyrosine_phosphatase-like
IPR036869 J_dom_sf
IPR010569 Myotubularin-like_Pase_dom
IPR001623 DnaJ_domain
SUPFAM
CDD
ProteinModelPortal
A0A194QLY7
A0A212F9V4
H9JCB3
A0A2H1VUC4
A0A1Y1KF83
A0A2J7PWP0
+ More
A0A067RKC6 A0A0P4VM95 T1HML4 A0A023F599 D6W9A3 A0A195BWH8 A0A154PFT4 E2B101 A0A195D9L4 A0A0L7RF14 A0A151WST9 A0A195CQ30 A0A195FQL7 A0A2R7VZ13 A0A3L8D828 A0A2A3EGH2 E0VQ65 E9IDA9 A0A2J7PWM9 A0A1S4EPX6 A0A2S2Q897 T1JEU2 J9K3U7 A0A310SPJ0 A0A2H8TFV5 A0A2P8XVN6 A0A1L8DMC6 B0W5Z6 C3Y1V6 A0A1Q3FDS2 A0A146P539 A0A3P8UT11 A0A3Q2FWL0 A0A2U9B189 A0A1L8I323 Q6AZN6 A0A3B4WVD8 A0A3B4TW17 G3Q5W9 A0A1A9WPA1 A0A3Q2PAN6 A0A3Q1FZN2 A0A3P8S0F3 A0A1W4ZW98 H2LUA0 A0A3P9LW04 A0A1B0CTU6 A0A1W4ZNA0 A0A1U8CKK8 A0A2I4CLW9 A0A3B3C9C2 A0A3Q2E6U0 A0A3Q3AKW4 G3Q5X0 Q6PF93 A0A3B5Q9A0 A0A1V4KTV3 I3K560 A0A3B3WQT0 A0A1U7RYK9 A0A3B3W1B4 A0A3S2PT87 A0A1J1ITM6 A0A147ASV9 A0A1A8RSW8 G1KPF8 F1Q9F3 A0A1A8NAL0 A0A1A8I9L5 A0A1A8UHN0 G1SX90 A0A3P9IUA7 A0A3Q3KSY3 A0A1A8C650 A0A146Y7U9 A0A3B4X232 A0A151MWA8 A0A3B1J562 A0A3Q2UEL4 A0A1S3SDV9 A0A3B4EA32 T1E4S4 A0A402EVR1 A0A1A8H9A0 A0A0F8B087 A0A060WR34 A0A3B3HSF3 A0A3P9KBE9 A0A1S3SDV8 J3SCI6 A0A3B4F817 A0A2I4CLX1 A0A3Q2VFU5 K9IP63
A0A067RKC6 A0A0P4VM95 T1HML4 A0A023F599 D6W9A3 A0A195BWH8 A0A154PFT4 E2B101 A0A195D9L4 A0A0L7RF14 A0A151WST9 A0A195CQ30 A0A195FQL7 A0A2R7VZ13 A0A3L8D828 A0A2A3EGH2 E0VQ65 E9IDA9 A0A2J7PWM9 A0A1S4EPX6 A0A2S2Q897 T1JEU2 J9K3U7 A0A310SPJ0 A0A2H8TFV5 A0A2P8XVN6 A0A1L8DMC6 B0W5Z6 C3Y1V6 A0A1Q3FDS2 A0A146P539 A0A3P8UT11 A0A3Q2FWL0 A0A2U9B189 A0A1L8I323 Q6AZN6 A0A3B4WVD8 A0A3B4TW17 G3Q5W9 A0A1A9WPA1 A0A3Q2PAN6 A0A3Q1FZN2 A0A3P8S0F3 A0A1W4ZW98 H2LUA0 A0A3P9LW04 A0A1B0CTU6 A0A1W4ZNA0 A0A1U8CKK8 A0A2I4CLW9 A0A3B3C9C2 A0A3Q2E6U0 A0A3Q3AKW4 G3Q5X0 Q6PF93 A0A3B5Q9A0 A0A1V4KTV3 I3K560 A0A3B3WQT0 A0A1U7RYK9 A0A3B3W1B4 A0A3S2PT87 A0A1J1ITM6 A0A147ASV9 A0A1A8RSW8 G1KPF8 F1Q9F3 A0A1A8NAL0 A0A1A8I9L5 A0A1A8UHN0 G1SX90 A0A3P9IUA7 A0A3Q3KSY3 A0A1A8C650 A0A146Y7U9 A0A3B4X232 A0A151MWA8 A0A3B1J562 A0A3Q2UEL4 A0A1S3SDV9 A0A3B4EA32 T1E4S4 A0A402EVR1 A0A1A8H9A0 A0A0F8B087 A0A060WR34 A0A3B3HSF3 A0A3P9KBE9 A0A1S3SDV8 J3SCI6 A0A3B4F817 A0A2I4CLX1 A0A3Q2VFU5 K9IP63
PDB
3LS8
E-value=0,
Score=1706
Ontologies
PATHWAY
00562
Inositol phosphate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04070 Phosphatidylinositol signaling system - Bombyx mori (domestic silkworm)
04136 Autophagy - other - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04145 Phagosome - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04070 Phosphatidylinositol signaling system - Bombyx mori (domestic silkworm)
04136 Autophagy - other - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04145 Phagosome - Bombyx mori (domestic silkworm)
GO
GO:0005524
GO:0016303
GO:0048015
GO:0036092
GO:0005737
GO:0006468
GO:0000407
GO:0006897
GO:0034272
GO:0000045
GO:0005768
GO:0005777
GO:0034271
GO:0030242
GO:0046854
GO:0016020
GO:0006261
GO:0003677
GO:0003887
GO:0016301
GO:0030496
GO:0032465
GO:0007049
GO:0051301
GO:0006914
GO:0016021
GO:0045335
GO:0004672
GO:0016236
GO:0016485
GO:0043201
GO:0045022
GO:0005829
GO:0035004
GO:0042149
GO:0035032
GO:0009267
GO:0044754
GO:0034497
GO:0050708
GO:0006497
GO:0005770
GO:0007032
GO:0005515
GO:0005488
GO:0004003
GO:0005634
GO:0006289
GO:0016286
PANTHER
Topology
Subcellular location
Midbody
Late endosome As component of the PI3K complex I localized to pre-autophagosome structures. As component of the PI3K complex II localized predominantly to endosomes (By similarity). Localizes also to discrete punctae along the ciliary axoneme and to the base of the ciliary axoneme (PubMed:24089209). With evidence from 3 publications.
Cytoplasmic vesicle As component of the PI3K complex I localized to pre-autophagosome structures. As component of the PI3K complex II localized predominantly to endosomes (By similarity). Localizes also to discrete punctae along the ciliary axoneme and to the base of the ciliary axoneme (PubMed:24089209). With evidence from 3 publications.
Autophagosome As component of the PI3K complex I localized to pre-autophagosome structures. As component of the PI3K complex II localized predominantly to endosomes (By similarity). Localizes also to discrete punctae along the ciliary axoneme and to the base of the ciliary axoneme (PubMed:24089209). With evidence from 3 publications.
Late endosome As component of the PI3K complex I localized to pre-autophagosome structures. As component of the PI3K complex II localized predominantly to endosomes (By similarity). Localizes also to discrete punctae along the ciliary axoneme and to the base of the ciliary axoneme (PubMed:24089209). With evidence from 3 publications.
Cytoplasmic vesicle As component of the PI3K complex I localized to pre-autophagosome structures. As component of the PI3K complex II localized predominantly to endosomes (By similarity). Localizes also to discrete punctae along the ciliary axoneme and to the base of the ciliary axoneme (PubMed:24089209). With evidence from 3 publications.
Autophagosome As component of the PI3K complex I localized to pre-autophagosome structures. As component of the PI3K complex II localized predominantly to endosomes (By similarity). Localizes also to discrete punctae along the ciliary axoneme and to the base of the ciliary axoneme (PubMed:24089209). With evidence from 3 publications.
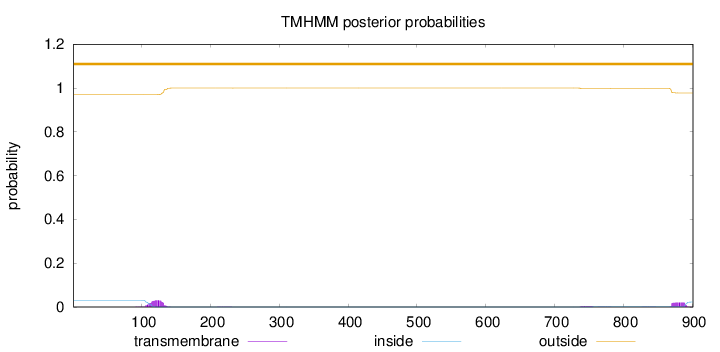
Length:
901
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.15403
Exp number, first 60 AAs:
5e-05
Total prob of N-in:
0.03043
outside
1 - 901
Population Genetic Test Statistics
Pi
22.718261
Theta
2.259455
Tajima's D
0.230657
CLR
0.938198
CSRT
0.480525973701315
Interpretation
Uncertain
