Gene
KWMTBOMO12763
Pre Gene Modal
BGIBMGA007200
Annotation
Vacuolar_protein_sorting_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 1.185 PlasmaMembrane Reliability : 1.288
Sequence
CDS
ATGGATCAACCACTCAATCTTAAATTGTCATCATTATCACTAATTTCACAAAACAAATTACAAAATATATTGAGTCAGTGTGGCGAGAAAAAGGATTTAATAATCGATCCATCCTTAATTAAGGCTTTGGAAAGAATATGCGGTGTGACATGGCTCAGACAACATGGAGTTGACAAAATATACAAAATGGATCCCCAACTCGGTCCAACGGCAAACGCAAATAGAGTATACTTTATTCCAGCGTGCATCATAAAATACAAATGTGTCCTGGACCAAATATCATCGCTCATTAGTCAGAATCCATCTATAGCTGACGTAAATGGCTTCCACATCATTATAATTCCAAAAGTCTTAAATTCATTTGATTCCATATTAGAAAGTAAAGGTTTATATGGTGTGGTTAAATTACATGCATTCGCATGGGAATTAATGGTCCTCGATGATCAGTTATTGAGTTTAGAGTTACCATTTTTATTTAAACAGTTATTTGTTGACCAAAATCAGTCCTTACTGTCCGGTGTAGCAATGTCTTTATGGACATTATTTCATGTGACAGGGAGACCTAAAGTAATGTGTTCATTTGGTAAATTTTCTGCGGGAGTACTAGACATGTTAGAAATATATAATGAAACTTATTCTAGAAACTTTGTAGATACAAATAATTCAAGGGATATAGGAGCGTTACTTGTTATAGATCGTACCCAGGATTATGCGTCAAGTCTGCTAACTCCAGCTACATACAGCGGATTGCTTAAAGAAATATTCAGTATAAACTGTGGATTTCTGGATTTGAATGTTAAAGATACAAAAGTAATGAAAGGCAAGTTAAATTTTGATAGTGAACAAAGCAATGAAAGTAAAAGTACAGCAATGCCTTTAGATAGTTCTATAGATAGTCTGTATGCTGAGATAAAACACAGACATTTCTCTGAAGTCCTCAGCGTTCTCAGTTCTAAAGCTAAATTACTTAAAAACGAAGACATAAAAGCATTAGGTATACAAGAAATGAAGCACTTTGTTGCGACCAAACTCCAACAAGTGACTCTCTTCAAGCAAAATCTAGTAAACCATGTGTTAGCTTGTGAAACGATAATATCAGAAATGAGCAACAGATTTGAGAATCTAAAATTAACTGAGACTGAAATGTTGAATAATAGAAACAAAAAGACCAATTATAACTTCCTTGATGAAAACTTCGGGACTGAAATTCACATTTATAATAGTTTGCGTCTGATGTGTTTGTTAAGCTTAACACAAGGTTTGTCTTATGATGAATACAATACATTAGTCAATAAATACCTCTTGGCCTTTGGTTACAAATATTTGTATGTATTCAATAATTTGATCAACTCTGGCTTGCTAATTCAACCTTCGAATATAAAGCTCTCATTAAATATTTCCAATTTAAGTAACTTAAGTGATAAATTACCAAAATGGCAGAATGAATTTCAGACTGCTGCTCATAAATTAAAACAGTTACCATCTAAGCCAGATGATGTAGGTAAGTCATCACCGAGTTATGTTTTCAACGGTGGCTACATACCTTTAGTTGCTATTATCTGTAATGCCATACTCACCTCTGAAAGCCTCAGTGAAGTTCTGACAAAGCTGTCAACTTTGACGGACTTTAAAATTGGTGGTGTAATAAATGAAAAACTGAAAGATGGTGTCGAGACGTTGAATGAGAAATTGTCAAATCTTAAAGTTTCAAATGACTTAGAATTTGGCTGCAAAGATCCTAAGATTTTATCTAAAGTAGTTAAAAGCGACCCGAACTTCTGTAATCTGTTTCAATTAAAGCCAAAAACTATTTTGGTATTTGTAATTGGAGGAGTGAACTATGCAGAGATAGCTGCATGTGAGGTTGTACAAACAAACACTGGAAGCAAGATTTATATTGCAAGTGATTGTGTATCAAATGGCAGTGATTTGATTGCTGCTCACATGTAA
Protein
MDQPLNLKLSSLSLISQNKLQNILSQCGEKKDLIIDPSLIKALERICGVTWLRQHGVDKIYKMDPQLGPTANANRVYFIPACIIKYKCVLDQISSLISQNPSIADVNGFHIIIIPKVLNSFDSILESKGLYGVVKLHAFAWELMVLDDQLLSLELPFLFKQLFVDQNQSLLSGVAMSLWTLFHVTGRPKVMCSFGKFSAGVLDMLEIYNETYSRNFVDTNNSRDIGALLVIDRTQDYASSLLTPATYSGLLKEIFSINCGFLDLNVKDTKVMKGKLNFDSEQSNESKSTAMPLDSSIDSLYAEIKHRHFSEVLSVLSSKAKLLKNEDIKALGIQEMKHFVATKLQQVTLFKQNLVNHVLACETIISEMSNRFENLKLTETEMLNNRNKKTNYNFLDENFGTEIHIYNSLRLMCLLSLTQGLSYDEYNTLVNKYLLAFGYKYLYVFNNLINSGLLIQPSNIKLSLNISNLSNLSDKLPKWQNEFQTAAHKLKQLPSKPDDVGKSSPSYVFNGGYIPLVAIICNAILTSESLSEVLTKLSTLTDFKIGGVINEKLKDGVETLNEKLSNLKVSNDLEFGCKDPKILSKVVKSDPNFCNLFQLKPKTILVFVIGGVNYAEIAACEVVQTNTGSKIYIASDCVSNGSDLIAAHM
Summary
Similarity
Belongs to the STXBP/unc-18/SEC1 family.
Uniprot
A0A0L7LAL5
A0A2A4JMJ8
A0A2W1BN30
A0A2H1W3D5
A0A212F9U4
A0A437B540
+ More
A0A0N1PG43 A0A194QHQ0 A0A1E1WJ48 A0A336MNX7 Q17EV6 A0A182RF24 A0A182JCX1 B0WYU9 A0A182QKT9 A0A182GW65 A0A1Q3F7K3 U5EYL5 A0A182VVQ8 A0A182NA07 A0A1Y1KUW7 A0A182JRL1 A0A1B0CSR8 A0A1W4XPI2 A0A182PH21 A0A182L9A8 A0A182HKY7 A0A182FSS0 A0A1J1HY38 Q7PUM3 A0A182UGZ2 A0A182VML1 A0A1I8MVJ0 W5J769 A0A182XA08 A0A0M3QY62 A0A067R5H4 A0A2J7RS11 T1I600 B4M469 A0A1B6FKL4 D6WB77 A0A1W4V9C5 Q294G5 A0A1B6JZG3 A0A0A9YVF1 B4KBV9 A0A0Q9XAE1 A0A3B0JP47 A0A1B6DTV4 N6U2R9 A0A146KXN7 A0A1A9VN03 A0A1B0A2I8 U4U269 A0A0L0CL10 A0A182M0U8 A0A2R7W001 A0A1B6LIA7 A0A2P8ZD26 A0A182G0Z2 A0A0A1WSQ6 A0A182YBB5 B4JFQ4 A0A1B0FD79 A0A1A9YEL4 W8C539 A0A034WDT1 A0A2J7RRZ5 B4QUW3 B4PTK7 B3P6J4 B4IC96 Q9VBR1 A0A2M4AR55 A0A2M4AQR8 K7J4Z4 A0A232EYV1 B3LW38 B4N864 A0A1B0DLC0 A0A2J7RRY5 A0A084W267 A0A2J7RRY4 A0A3B0KJR5 A0A1I8NSV3 A0A182SHP5 A0A0K8VWE5 A0A195C472 V3ZES9 A0A026WS41 A0A2A3EER6 A0A087ZSK7 Q8SZS6 A0A310SFI5 E2BTZ9 A0A195BUQ8
A0A0N1PG43 A0A194QHQ0 A0A1E1WJ48 A0A336MNX7 Q17EV6 A0A182RF24 A0A182JCX1 B0WYU9 A0A182QKT9 A0A182GW65 A0A1Q3F7K3 U5EYL5 A0A182VVQ8 A0A182NA07 A0A1Y1KUW7 A0A182JRL1 A0A1B0CSR8 A0A1W4XPI2 A0A182PH21 A0A182L9A8 A0A182HKY7 A0A182FSS0 A0A1J1HY38 Q7PUM3 A0A182UGZ2 A0A182VML1 A0A1I8MVJ0 W5J769 A0A182XA08 A0A0M3QY62 A0A067R5H4 A0A2J7RS11 T1I600 B4M469 A0A1B6FKL4 D6WB77 A0A1W4V9C5 Q294G5 A0A1B6JZG3 A0A0A9YVF1 B4KBV9 A0A0Q9XAE1 A0A3B0JP47 A0A1B6DTV4 N6U2R9 A0A146KXN7 A0A1A9VN03 A0A1B0A2I8 U4U269 A0A0L0CL10 A0A182M0U8 A0A2R7W001 A0A1B6LIA7 A0A2P8ZD26 A0A182G0Z2 A0A0A1WSQ6 A0A182YBB5 B4JFQ4 A0A1B0FD79 A0A1A9YEL4 W8C539 A0A034WDT1 A0A2J7RRZ5 B4QUW3 B4PTK7 B3P6J4 B4IC96 Q9VBR1 A0A2M4AR55 A0A2M4AQR8 K7J4Z4 A0A232EYV1 B3LW38 B4N864 A0A1B0DLC0 A0A2J7RRY5 A0A084W267 A0A2J7RRY4 A0A3B0KJR5 A0A1I8NSV3 A0A182SHP5 A0A0K8VWE5 A0A195C472 V3ZES9 A0A026WS41 A0A2A3EER6 A0A087ZSK7 Q8SZS6 A0A310SFI5 E2BTZ9 A0A195BUQ8
Pubmed
26227816
28756777
22118469
26354079
17510324
26483478
+ More
28004739 20966253 12364791 14747013 17210077 25315136 20920257 23761445 24845553 17994087 18362917 19820115 15632085 25401762 23537049 26823975 26108605 29403074 25830018 25244985 24495485 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20075255 28648823 24438588 23254933 24508170 30249741 20798317
28004739 20966253 12364791 14747013 17210077 25315136 20920257 23761445 24845553 17994087 18362917 19820115 15632085 25401762 23537049 26823975 26108605 29403074 25830018 25244985 24495485 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20075255 28648823 24438588 23254933 24508170 30249741 20798317
EMBL
JTDY01001960
KOB72444.1
NWSH01001097
PCG72620.1
KZ150013
PZC75034.1
+ More
ODYU01005928 SOQ47332.1 AGBW02009556 OWR50515.1 RSAL01000164 RVE45333.1 KQ460882 KPJ11467.1 KQ458981 KPJ04465.1 GDQN01004096 JAT86958.1 UFQS01001942 UFQT01001942 SSX12752.1 SSX32194.1 CH477279 EAT44995.1 DS232199 EDS37209.1 AXCN02001521 JXUM01092595 KQ563997 KXJ72951.1 GFDL01011513 JAV23532.1 GANO01000222 JAB59649.1 GEZM01075772 JAV63960.1 AJWK01026493 AJWK01026494 APCN01000875 CVRI01000024 CRK92252.1 AAAB01008987 EAA01309.4 ADMH02002125 ETN58710.1 CP012526 ALC47123.1 KK852872 KDR14570.1 NEVH01000597 PNF43602.1 ACPB03008320 CH940652 EDW59430.1 GECZ01019049 JAS50720.1 KQ971312 EEZ98704.1 CM000070 EAL28999.3 GECU01003393 JAT04314.1 GBHO01010054 GBRD01015522 JAG33550.1 JAG50304.1 CH933806 EDW15811.1 KRG01746.1 OUUW01000008 SPP83997.1 GEDC01008192 JAS29106.1 APGK01044427 KB741026 ENN74911.1 GDHC01018264 GDHC01002937 JAQ00365.1 JAQ15692.1 KB631924 ERL87167.1 JRES01000254 KNC32936.1 AXCM01000128 KK854153 PTY12260.1 GEBQ01016633 JAT23344.1 PYGN01000098 PSN54370.1 JXUM01037701 KQ561138 KXJ79601.1 GBXI01012445 JAD01847.1 CH916369 EDV93535.1 CCAG010017532 GAMC01001987 JAC04569.1 GAKP01006103 JAC52849.1 PNF43604.1 CM000364 EDX14429.1 CM000160 EDW98747.1 CH954182 EDV53664.1 CH480828 EDW44992.1 AE014297 AAF56468.1 GGFK01009767 MBW43088.1 GGFK01009741 MBW43062.1 NNAY01001584 OXU23523.1 CH902617 EDV42616.1 CH964232 EDW81315.2 AJVK01071103 PNF43601.1 ATLV01019524 KE525273 KFB44311.1 PNF43603.1 SPP83998.1 GDHF01009125 JAI43189.1 KQ978344 KYM94976.1 KB202567 ESO89653.1 KK107119 QOIP01000009 EZA58768.1 RLU18858.1 KZ288266 PBC30210.1 AY070542 AAL48013.1 KQ760505 OAD60075.1 GL450531 EFN80898.1 KQ976406 KYM91657.1
ODYU01005928 SOQ47332.1 AGBW02009556 OWR50515.1 RSAL01000164 RVE45333.1 KQ460882 KPJ11467.1 KQ458981 KPJ04465.1 GDQN01004096 JAT86958.1 UFQS01001942 UFQT01001942 SSX12752.1 SSX32194.1 CH477279 EAT44995.1 DS232199 EDS37209.1 AXCN02001521 JXUM01092595 KQ563997 KXJ72951.1 GFDL01011513 JAV23532.1 GANO01000222 JAB59649.1 GEZM01075772 JAV63960.1 AJWK01026493 AJWK01026494 APCN01000875 CVRI01000024 CRK92252.1 AAAB01008987 EAA01309.4 ADMH02002125 ETN58710.1 CP012526 ALC47123.1 KK852872 KDR14570.1 NEVH01000597 PNF43602.1 ACPB03008320 CH940652 EDW59430.1 GECZ01019049 JAS50720.1 KQ971312 EEZ98704.1 CM000070 EAL28999.3 GECU01003393 JAT04314.1 GBHO01010054 GBRD01015522 JAG33550.1 JAG50304.1 CH933806 EDW15811.1 KRG01746.1 OUUW01000008 SPP83997.1 GEDC01008192 JAS29106.1 APGK01044427 KB741026 ENN74911.1 GDHC01018264 GDHC01002937 JAQ00365.1 JAQ15692.1 KB631924 ERL87167.1 JRES01000254 KNC32936.1 AXCM01000128 KK854153 PTY12260.1 GEBQ01016633 JAT23344.1 PYGN01000098 PSN54370.1 JXUM01037701 KQ561138 KXJ79601.1 GBXI01012445 JAD01847.1 CH916369 EDV93535.1 CCAG010017532 GAMC01001987 JAC04569.1 GAKP01006103 JAC52849.1 PNF43604.1 CM000364 EDX14429.1 CM000160 EDW98747.1 CH954182 EDV53664.1 CH480828 EDW44992.1 AE014297 AAF56468.1 GGFK01009767 MBW43088.1 GGFK01009741 MBW43062.1 NNAY01001584 OXU23523.1 CH902617 EDV42616.1 CH964232 EDW81315.2 AJVK01071103 PNF43601.1 ATLV01019524 KE525273 KFB44311.1 PNF43603.1 SPP83998.1 GDHF01009125 JAI43189.1 KQ978344 KYM94976.1 KB202567 ESO89653.1 KK107119 QOIP01000009 EZA58768.1 RLU18858.1 KZ288266 PBC30210.1 AY070542 AAL48013.1 KQ760505 OAD60075.1 GL450531 EFN80898.1 KQ976406 KYM91657.1
Proteomes
UP000037510
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000008820 UP000075900 UP000075880 UP000002320 UP000075886 UP000069940 UP000249989 UP000075920 UP000075884 UP000075881 UP000092461 UP000192223 UP000075885 UP000075882 UP000075840 UP000069272 UP000183832 UP000007062 UP000075902 UP000075903 UP000095301 UP000000673 UP000076407 UP000092553 UP000027135 UP000235965 UP000015103 UP000008792 UP000007266 UP000192221 UP000001819 UP000009192 UP000268350 UP000019118 UP000078200 UP000092445 UP000030742 UP000037069 UP000075883 UP000245037 UP000076408 UP000001070 UP000092444 UP000092443 UP000000304 UP000002282 UP000008711 UP000001292 UP000000803 UP000002358 UP000215335 UP000007801 UP000007798 UP000092462 UP000030765 UP000095300 UP000075901 UP000078542 UP000030746 UP000053097 UP000279307 UP000242457 UP000005203 UP000008237 UP000078540
UP000008820 UP000075900 UP000075880 UP000002320 UP000075886 UP000069940 UP000249989 UP000075920 UP000075884 UP000075881 UP000092461 UP000192223 UP000075885 UP000075882 UP000075840 UP000069272 UP000183832 UP000007062 UP000075902 UP000075903 UP000095301 UP000000673 UP000076407 UP000092553 UP000027135 UP000235965 UP000015103 UP000008792 UP000007266 UP000192221 UP000001819 UP000009192 UP000268350 UP000019118 UP000078200 UP000092445 UP000030742 UP000037069 UP000075883 UP000245037 UP000076408 UP000001070 UP000092444 UP000092443 UP000000304 UP000002282 UP000008711 UP000001292 UP000000803 UP000002358 UP000215335 UP000007801 UP000007798 UP000092462 UP000030765 UP000095300 UP000075901 UP000078542 UP000030746 UP000053097 UP000279307 UP000242457 UP000005203 UP000008237 UP000078540
Interpro
Gene 3D
ProteinModelPortal
A0A0L7LAL5
A0A2A4JMJ8
A0A2W1BN30
A0A2H1W3D5
A0A212F9U4
A0A437B540
+ More
A0A0N1PG43 A0A194QHQ0 A0A1E1WJ48 A0A336MNX7 Q17EV6 A0A182RF24 A0A182JCX1 B0WYU9 A0A182QKT9 A0A182GW65 A0A1Q3F7K3 U5EYL5 A0A182VVQ8 A0A182NA07 A0A1Y1KUW7 A0A182JRL1 A0A1B0CSR8 A0A1W4XPI2 A0A182PH21 A0A182L9A8 A0A182HKY7 A0A182FSS0 A0A1J1HY38 Q7PUM3 A0A182UGZ2 A0A182VML1 A0A1I8MVJ0 W5J769 A0A182XA08 A0A0M3QY62 A0A067R5H4 A0A2J7RS11 T1I600 B4M469 A0A1B6FKL4 D6WB77 A0A1W4V9C5 Q294G5 A0A1B6JZG3 A0A0A9YVF1 B4KBV9 A0A0Q9XAE1 A0A3B0JP47 A0A1B6DTV4 N6U2R9 A0A146KXN7 A0A1A9VN03 A0A1B0A2I8 U4U269 A0A0L0CL10 A0A182M0U8 A0A2R7W001 A0A1B6LIA7 A0A2P8ZD26 A0A182G0Z2 A0A0A1WSQ6 A0A182YBB5 B4JFQ4 A0A1B0FD79 A0A1A9YEL4 W8C539 A0A034WDT1 A0A2J7RRZ5 B4QUW3 B4PTK7 B3P6J4 B4IC96 Q9VBR1 A0A2M4AR55 A0A2M4AQR8 K7J4Z4 A0A232EYV1 B3LW38 B4N864 A0A1B0DLC0 A0A2J7RRY5 A0A084W267 A0A2J7RRY4 A0A3B0KJR5 A0A1I8NSV3 A0A182SHP5 A0A0K8VWE5 A0A195C472 V3ZES9 A0A026WS41 A0A2A3EER6 A0A087ZSK7 Q8SZS6 A0A310SFI5 E2BTZ9 A0A195BUQ8
A0A0N1PG43 A0A194QHQ0 A0A1E1WJ48 A0A336MNX7 Q17EV6 A0A182RF24 A0A182JCX1 B0WYU9 A0A182QKT9 A0A182GW65 A0A1Q3F7K3 U5EYL5 A0A182VVQ8 A0A182NA07 A0A1Y1KUW7 A0A182JRL1 A0A1B0CSR8 A0A1W4XPI2 A0A182PH21 A0A182L9A8 A0A182HKY7 A0A182FSS0 A0A1J1HY38 Q7PUM3 A0A182UGZ2 A0A182VML1 A0A1I8MVJ0 W5J769 A0A182XA08 A0A0M3QY62 A0A067R5H4 A0A2J7RS11 T1I600 B4M469 A0A1B6FKL4 D6WB77 A0A1W4V9C5 Q294G5 A0A1B6JZG3 A0A0A9YVF1 B4KBV9 A0A0Q9XAE1 A0A3B0JP47 A0A1B6DTV4 N6U2R9 A0A146KXN7 A0A1A9VN03 A0A1B0A2I8 U4U269 A0A0L0CL10 A0A182M0U8 A0A2R7W001 A0A1B6LIA7 A0A2P8ZD26 A0A182G0Z2 A0A0A1WSQ6 A0A182YBB5 B4JFQ4 A0A1B0FD79 A0A1A9YEL4 W8C539 A0A034WDT1 A0A2J7RRZ5 B4QUW3 B4PTK7 B3P6J4 B4IC96 Q9VBR1 A0A2M4AR55 A0A2M4AQR8 K7J4Z4 A0A232EYV1 B3LW38 B4N864 A0A1B0DLC0 A0A2J7RRY5 A0A084W267 A0A2J7RRY4 A0A3B0KJR5 A0A1I8NSV3 A0A182SHP5 A0A0K8VWE5 A0A195C472 V3ZES9 A0A026WS41 A0A2A3EER6 A0A087ZSK7 Q8SZS6 A0A310SFI5 E2BTZ9 A0A195BUQ8
PDB
4BX9
E-value=1.84412e-23,
Score=272
Ontologies
GO
PANTHER
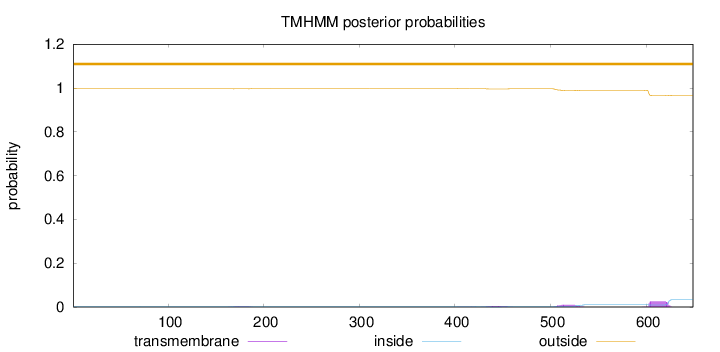
Topology
Length:
649
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.906819999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00309
outside
1 - 649
Population Genetic Test Statistics
Pi
25.39395
Theta
20.799785
Tajima's D
0.650844
CLR
0.647313
CSRT
0.568871556422179
Interpretation
Uncertain
