Pre Gene Modal
BGIBMGA007160
Annotation
PREDICTED:_probable_isocitrate_dehydrogenase_[NAD]_subunit_alpha?_mitochondrial_[Bombyx_mori]
Full name
Isocitrate dehydrogenase [NAD] subunit, mitochondrial
Location in the cell
Cytoplasmic Reliability : 1.958
Sequence
CDS
ATGTCAGACGGCCTCTTCCTGCGCTGTTGTCGCGAGCTCGCCACCAAGTACCCGGACATCAAGTTTGAAGAGCGATACCTGGACACGGTCTGCCTCAACATGGTGCAGGACCCCTCCAAGTTTGACGTGCTGGTGATGCCCAACCTGTACGGTGACATCATGTCGGACATGTGTTCCGGGCTCGTGGGCGGGCTCGGCCTCACGCCCTCCGGCAACATCGGCAAGAACGGCGCGCTCTTCGAGTCGGTACACGGTACAGCCCCAGATATCGCCGGTAAAGACATGGCCAATCCTACAGCCCTACTGTTGTCGGCTATTATGATGCTGAGGCATTTGCAGTTAAATGATCACGCTGATCTCGTCCAGAATGCCTGTTACGAAGTATTACGCGAGGGCAGAGCCTTGACTGGTGACCTCGGCGGTACCGCGAAATGCAGTGAATTTACTAATGCCATCATTTCAAAACTGAATTAG
Protein
MSDGLFLRCCRELATKYPDIKFEERYLDTVCLNMVQDPSKFDVLVMPNLYGDIMSDMCSGLVGGLGLTPSGNIGKNGALFESVHGTAPDIAGKDMANPTALLLSAIMMLRHLQLNDHADLVQNACYEVLREGRALTGDLGGTAKCSEFTNAIISKLN
Summary
Similarity
Belongs to the isocitrate and isopropylmalate dehydrogenases family.
Uniprot
I4DNF6
A0A3S2L4B0
S4PM50
A0A1S4EVW3
Q17P80
A0A023EQF0
+ More
Q17P79 A0A1S4EVV7 A0A182SRU0 H9JCB5 A0A1A9W960 T1D494 T1D2I5 A0A1B0FFJ0 A0A182JHS8 A0A034VLP0 A0A084VRI3 A0A034VRS1 D3TSB1 A0A182QRH2 A0A1A9URI5 A0A0A1XLS9 A0A2M4AER3 A0A182M3T4 A0A0A1WTA0 A0A1A9YJC5 A0A2M4AC44 A0A2M4BSD8 W5JJ40 A0A182TG30 A0A1I8JVG7 A0A340TB72 A0A182LE36 A0A2C9GLB7 Q7PQX9 T1DQ50 A0A2M4BSU9 A0A182FQY2 A0A182RT14 A0A1Y9HD80 B0W6Q6 A0A2M4BQQ7 A0A182NR46 A0A2M4ADC4 A0A453YML4 F5HKN6 A0A182UYU4 A0A182HS68 A0A1Q3G3W3 A0A1Q3FGV2 A0A182PCK6 A0A2M3Z5S3 A0A182XAC4 W8CDU6 A0A182VZT2 A0A2M4ANN7 A0A2M4CFZ9 A0A182U9D6 A0A1B6GXP0 A0A182Y3Z3 A0A2M3Z5N9 A0A182K4P9 A0A1B0EZB5 A0A0L0C9V8 A0A1L8E0K4 J3JXM4 A0A1L8E022 A0A1Q3G3X4 A0A0K8TTP8 A0A1J1J2D3 A0A0N0PBV0 A0A1L8E026 T1P9X3 U5EUV7 A0A212F9U9 A0A1I8P883 D6WCZ6 A0A2H1W2T3 A0A0M4ETP7 A0A0C5QEU6 A0A1L8EDB1 A0A1L8EHZ0 A0A1W4W9H9 A0A336JXZ0 J9JQ96 B4JX16 B4L6W2 A0A1S3DCF1 A0A0Q9XF11 A0A0K2VI16 E2B105 A0A154PEB2 A0A1B6KN33 A0A3B0KVC3 A0A2S2NBN6 A0A1W4VG47 A0A2W1BIY5 A0A0P8Y537 A0A310SK85 A0A1W4VTF1
Q17P79 A0A1S4EVV7 A0A182SRU0 H9JCB5 A0A1A9W960 T1D494 T1D2I5 A0A1B0FFJ0 A0A182JHS8 A0A034VLP0 A0A084VRI3 A0A034VRS1 D3TSB1 A0A182QRH2 A0A1A9URI5 A0A0A1XLS9 A0A2M4AER3 A0A182M3T4 A0A0A1WTA0 A0A1A9YJC5 A0A2M4AC44 A0A2M4BSD8 W5JJ40 A0A182TG30 A0A1I8JVG7 A0A340TB72 A0A182LE36 A0A2C9GLB7 Q7PQX9 T1DQ50 A0A2M4BSU9 A0A182FQY2 A0A182RT14 A0A1Y9HD80 B0W6Q6 A0A2M4BQQ7 A0A182NR46 A0A2M4ADC4 A0A453YML4 F5HKN6 A0A182UYU4 A0A182HS68 A0A1Q3G3W3 A0A1Q3FGV2 A0A182PCK6 A0A2M3Z5S3 A0A182XAC4 W8CDU6 A0A182VZT2 A0A2M4ANN7 A0A2M4CFZ9 A0A182U9D6 A0A1B6GXP0 A0A182Y3Z3 A0A2M3Z5N9 A0A182K4P9 A0A1B0EZB5 A0A0L0C9V8 A0A1L8E0K4 J3JXM4 A0A1L8E022 A0A1Q3G3X4 A0A0K8TTP8 A0A1J1J2D3 A0A0N0PBV0 A0A1L8E026 T1P9X3 U5EUV7 A0A212F9U9 A0A1I8P883 D6WCZ6 A0A2H1W2T3 A0A0M4ETP7 A0A0C5QEU6 A0A1L8EDB1 A0A1L8EHZ0 A0A1W4W9H9 A0A336JXZ0 J9JQ96 B4JX16 B4L6W2 A0A1S3DCF1 A0A0Q9XF11 A0A0K2VI16 E2B105 A0A154PEB2 A0A1B6KN33 A0A3B0KVC3 A0A2S2NBN6 A0A1W4VG47 A0A2W1BIY5 A0A0P8Y537 A0A310SK85 A0A1W4VTF1
Pubmed
EMBL
AK402883
BAM19446.1
RSAL01000164
RVE45334.1
GAIX01003740
JAA88820.1
+ More
CH477193 EAT48554.1 GAPW01002275 JAC11323.1 EAT48553.1 BABH01031587 GALA01001043 JAA93809.1 GALA01001603 JAA93249.1 CCAG010015760 GAKP01014731 JAC44221.1 ATLV01015680 KE525027 KFB40577.1 GAKP01014729 JAC44223.1 EZ424313 ADD20589.1 AXCN02001476 GBXI01002502 JAD11790.1 GGFK01005891 MBW39212.1 AXCM01000347 GBXI01012205 JAD02087.1 GGFK01004971 MBW38292.1 GGFJ01006829 MBW55970.1 ADMH02001265 ETN63313.1 APCN01000245 AAAB01008859 EAA08136.6 GAMD01001385 JAB00206.1 GGFJ01006998 MBW56139.1 DS231849 EDS36887.1 GGFJ01006275 MBW55416.1 GGFK01005419 MBW38740.1 EGK96788.1 GFDL01000550 JAV34495.1 GFDL01008248 JAV26797.1 GGFM01003120 MBW23871.1 GAMC01001331 JAC05225.1 GGFK01009079 MBW42400.1 GGFL01000001 MBW64179.1 GECZ01002573 JAS67196.1 GGFM01003079 MBW23830.1 AJVK01008650 JRES01000713 KNC29037.1 GFDF01001982 JAV12102.1 APGK01052691 BT127994 KB741213 KB631874 AEE62956.1 ENN72661.1 ERL86877.1 GFDF01001991 JAV12093.1 GFDL01000537 JAV34508.1 GDAI01000080 JAI17523.1 CVRI01000067 CRL06699.1 KQ460882 KPJ11466.1 GFDF01001974 JAV12110.1 KA645474 AFP60103.1 GANO01001283 JAB58588.1 AGBW02009556 OWR50516.1 KQ971311 EEZ99082.2 ODYU01005928 SOQ47333.1 CP012528 ALC49940.1 KM609498 AJQ30122.1 GFDG01002157 JAV16642.1 GFDG01000580 JAV18219.1 UFQS01000006 UFQT01000006 SSW97007.1 SSX17394.1 ABLF02036961 ABLF02036966 CH916376 EDV95292.1 CH933812 EDW06108.1 KRG07151.1 HACA01032718 CDW50079.1 GL444767 EFN60631.1 KQ434886 KZC10183.1 GEBQ01027125 JAT12852.1 OUUW01000031 SPP89806.1 GGMR01001994 MBY14613.1 KZ150013 PZC75032.1 CH902621 KPU81686.1 KQ762031 OAD56356.1
CH477193 EAT48554.1 GAPW01002275 JAC11323.1 EAT48553.1 BABH01031587 GALA01001043 JAA93809.1 GALA01001603 JAA93249.1 CCAG010015760 GAKP01014731 JAC44221.1 ATLV01015680 KE525027 KFB40577.1 GAKP01014729 JAC44223.1 EZ424313 ADD20589.1 AXCN02001476 GBXI01002502 JAD11790.1 GGFK01005891 MBW39212.1 AXCM01000347 GBXI01012205 JAD02087.1 GGFK01004971 MBW38292.1 GGFJ01006829 MBW55970.1 ADMH02001265 ETN63313.1 APCN01000245 AAAB01008859 EAA08136.6 GAMD01001385 JAB00206.1 GGFJ01006998 MBW56139.1 DS231849 EDS36887.1 GGFJ01006275 MBW55416.1 GGFK01005419 MBW38740.1 EGK96788.1 GFDL01000550 JAV34495.1 GFDL01008248 JAV26797.1 GGFM01003120 MBW23871.1 GAMC01001331 JAC05225.1 GGFK01009079 MBW42400.1 GGFL01000001 MBW64179.1 GECZ01002573 JAS67196.1 GGFM01003079 MBW23830.1 AJVK01008650 JRES01000713 KNC29037.1 GFDF01001982 JAV12102.1 APGK01052691 BT127994 KB741213 KB631874 AEE62956.1 ENN72661.1 ERL86877.1 GFDF01001991 JAV12093.1 GFDL01000537 JAV34508.1 GDAI01000080 JAI17523.1 CVRI01000067 CRL06699.1 KQ460882 KPJ11466.1 GFDF01001974 JAV12110.1 KA645474 AFP60103.1 GANO01001283 JAB58588.1 AGBW02009556 OWR50516.1 KQ971311 EEZ99082.2 ODYU01005928 SOQ47333.1 CP012528 ALC49940.1 KM609498 AJQ30122.1 GFDG01002157 JAV16642.1 GFDG01000580 JAV18219.1 UFQS01000006 UFQT01000006 SSW97007.1 SSX17394.1 ABLF02036961 ABLF02036966 CH916376 EDV95292.1 CH933812 EDW06108.1 KRG07151.1 HACA01032718 CDW50079.1 GL444767 EFN60631.1 KQ434886 KZC10183.1 GEBQ01027125 JAT12852.1 OUUW01000031 SPP89806.1 GGMR01001994 MBY14613.1 KZ150013 PZC75032.1 CH902621 KPU81686.1 KQ762031 OAD56356.1
Proteomes
UP000283053
UP000008820
UP000075901
UP000005204
UP000091820
UP000092444
+ More
UP000075880 UP000030765 UP000075886 UP000078200 UP000075883 UP000092443 UP000000673 UP000075902 UP000076407 UP000075903 UP000075882 UP000075840 UP000007062 UP000069272 UP000075900 UP000002320 UP000075884 UP000075885 UP000075920 UP000076408 UP000075881 UP000092462 UP000037069 UP000019118 UP000030742 UP000183832 UP000053240 UP000095301 UP000007151 UP000095300 UP000007266 UP000092553 UP000192223 UP000007819 UP000001070 UP000009192 UP000079169 UP000000311 UP000076502 UP000268350 UP000192221 UP000007801
UP000075880 UP000030765 UP000075886 UP000078200 UP000075883 UP000092443 UP000000673 UP000075902 UP000076407 UP000075903 UP000075882 UP000075840 UP000007062 UP000069272 UP000075900 UP000002320 UP000075884 UP000075885 UP000075920 UP000076408 UP000075881 UP000092462 UP000037069 UP000019118 UP000030742 UP000183832 UP000053240 UP000095301 UP000007151 UP000095300 UP000007266 UP000092553 UP000192223 UP000007819 UP000001070 UP000009192 UP000079169 UP000000311 UP000076502 UP000268350 UP000192221 UP000007801
PRIDE
Pfam
PF00180 Iso_dh
Interpro
ProteinModelPortal
I4DNF6
A0A3S2L4B0
S4PM50
A0A1S4EVW3
Q17P80
A0A023EQF0
+ More
Q17P79 A0A1S4EVV7 A0A182SRU0 H9JCB5 A0A1A9W960 T1D494 T1D2I5 A0A1B0FFJ0 A0A182JHS8 A0A034VLP0 A0A084VRI3 A0A034VRS1 D3TSB1 A0A182QRH2 A0A1A9URI5 A0A0A1XLS9 A0A2M4AER3 A0A182M3T4 A0A0A1WTA0 A0A1A9YJC5 A0A2M4AC44 A0A2M4BSD8 W5JJ40 A0A182TG30 A0A1I8JVG7 A0A340TB72 A0A182LE36 A0A2C9GLB7 Q7PQX9 T1DQ50 A0A2M4BSU9 A0A182FQY2 A0A182RT14 A0A1Y9HD80 B0W6Q6 A0A2M4BQQ7 A0A182NR46 A0A2M4ADC4 A0A453YML4 F5HKN6 A0A182UYU4 A0A182HS68 A0A1Q3G3W3 A0A1Q3FGV2 A0A182PCK6 A0A2M3Z5S3 A0A182XAC4 W8CDU6 A0A182VZT2 A0A2M4ANN7 A0A2M4CFZ9 A0A182U9D6 A0A1B6GXP0 A0A182Y3Z3 A0A2M3Z5N9 A0A182K4P9 A0A1B0EZB5 A0A0L0C9V8 A0A1L8E0K4 J3JXM4 A0A1L8E022 A0A1Q3G3X4 A0A0K8TTP8 A0A1J1J2D3 A0A0N0PBV0 A0A1L8E026 T1P9X3 U5EUV7 A0A212F9U9 A0A1I8P883 D6WCZ6 A0A2H1W2T3 A0A0M4ETP7 A0A0C5QEU6 A0A1L8EDB1 A0A1L8EHZ0 A0A1W4W9H9 A0A336JXZ0 J9JQ96 B4JX16 B4L6W2 A0A1S3DCF1 A0A0Q9XF11 A0A0K2VI16 E2B105 A0A154PEB2 A0A1B6KN33 A0A3B0KVC3 A0A2S2NBN6 A0A1W4VG47 A0A2W1BIY5 A0A0P8Y537 A0A310SK85 A0A1W4VTF1
Q17P79 A0A1S4EVV7 A0A182SRU0 H9JCB5 A0A1A9W960 T1D494 T1D2I5 A0A1B0FFJ0 A0A182JHS8 A0A034VLP0 A0A084VRI3 A0A034VRS1 D3TSB1 A0A182QRH2 A0A1A9URI5 A0A0A1XLS9 A0A2M4AER3 A0A182M3T4 A0A0A1WTA0 A0A1A9YJC5 A0A2M4AC44 A0A2M4BSD8 W5JJ40 A0A182TG30 A0A1I8JVG7 A0A340TB72 A0A182LE36 A0A2C9GLB7 Q7PQX9 T1DQ50 A0A2M4BSU9 A0A182FQY2 A0A182RT14 A0A1Y9HD80 B0W6Q6 A0A2M4BQQ7 A0A182NR46 A0A2M4ADC4 A0A453YML4 F5HKN6 A0A182UYU4 A0A182HS68 A0A1Q3G3W3 A0A1Q3FGV2 A0A182PCK6 A0A2M3Z5S3 A0A182XAC4 W8CDU6 A0A182VZT2 A0A2M4ANN7 A0A2M4CFZ9 A0A182U9D6 A0A1B6GXP0 A0A182Y3Z3 A0A2M3Z5N9 A0A182K4P9 A0A1B0EZB5 A0A0L0C9V8 A0A1L8E0K4 J3JXM4 A0A1L8E022 A0A1Q3G3X4 A0A0K8TTP8 A0A1J1J2D3 A0A0N0PBV0 A0A1L8E026 T1P9X3 U5EUV7 A0A212F9U9 A0A1I8P883 D6WCZ6 A0A2H1W2T3 A0A0M4ETP7 A0A0C5QEU6 A0A1L8EDB1 A0A1L8EHZ0 A0A1W4W9H9 A0A336JXZ0 J9JQ96 B4JX16 B4L6W2 A0A1S3DCF1 A0A0Q9XF11 A0A0K2VI16 E2B105 A0A154PEB2 A0A1B6KN33 A0A3B0KVC3 A0A2S2NBN6 A0A1W4VG47 A0A2W1BIY5 A0A0P8Y537 A0A310SK85 A0A1W4VTF1
PDB
5YVT
E-value=1.6113e-64,
Score=619
Ontologies
GO
Topology
Subcellular location
Mitochondrion
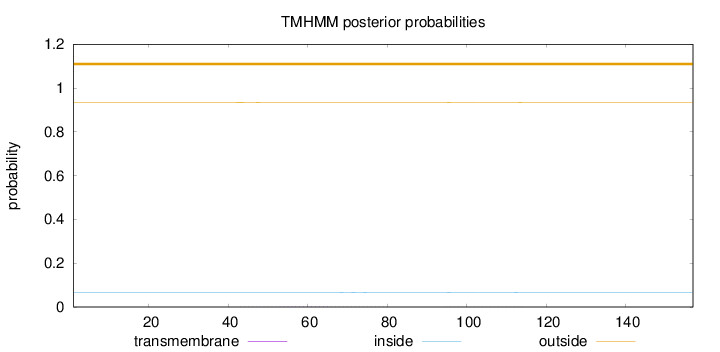
Length:
157
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04426
Exp number, first 60 AAs:
0.01754
Total prob of N-in:
0.06520
outside
1 - 157
Population Genetic Test Statistics
Pi
26.975232
Theta
27.502831
Tajima's D
0.138315
CLR
0.220482
CSRT
0.411629418529074
Interpretation
Uncertain
