Gene
KWMTBOMO12758
Pre Gene Modal
BGIBMGA007199
Annotation
PREDICTED:_uncharacterized_family_31_glucosidase_KIAA1161-like_[Amyelois_transitella]
Location in the cell
Lysosomal Reliability : 1.481
Sequence
CDS
ATGGACGCCAAAATATTCTTACTACTGTTATCGACAGTAGTATTTGTCAATGCTGTAGACATATCACTCATAGACTCGACTGACGTCAAATTGAAACTACAAGACAACCCATATGGAGGACTTAACGTACTTTTAAATAGAAGAGGCATTTCAGAAGTGATTTCAGTCATAGGACGTCGAACGTCTGGCATAAAGGACATAATTCAGAACGGCGACGTATTGATCCAGTTCAAAAACGACACCACGGTCAGAATCAGTGTCGAGCATATCGCGGGTGAGAGGAAAGGTCACCTGCTGTCGTTCGCTTGGAAAACAACAAAGAACACCCGACTGGAGGACTGTGCTAATCTCGGCACCGACAGCTGGTATGGAGGCCCGCAACAAAAACGCCAATACTGGCCCATCGAGAGACTCGTTCTCCCGAACTACTCGTATGTGACCAAGGAAGCGGACAACAGTGCTGTAGCCGAGCCTTACTGGCTCAACTCCCAGGGCATATTTTATTACTTCGACAAAAAAGTCCCATTATTCATTGACCAAAACGACTTAGAAAAGAATGCCGCTTGCTTCATAGCTCAGATCAGACCCCCGTATTCAAAGAATAGAGAGAGCAACGAACTCGTATACGCTTTAGGCATCTTCGGTGACGTCAAACAGGCCCATCAATTCGCAGTAGAGAAATATTTGAACAAGCCCAAAGCGATTCCTGATGAGCGAATGATAAAGTACCCTATATGGTCAACCTGGGCCAGGTACAAAAGGGACGTGAACCATGACGTTGTATTAAAATTCGCCGATGAAATAAATAAAAATGGTTTTCCTAATAGTCAAATGGAAATCGACGATCTATGGGAGATTTGTTATGGTTCCCAAACTGTTGATATTAGAAGATTTCCCGACATGAAAGATATGGTTGGAAAACTCAAAGCTAAAGGTTTCCGGGTGACGCTTTGGACCCATCCATTTATAAACAAAGGTTGCGAGCCATGGTATTCTGAAGCCAAAGCGAAGGGTTATTTAGTTTCTTCTGAATACGGTTCAGTGGAAACGAGCTGGTGGAACGACAATGGCACCACCACATCATACATTGATTTCACCAAACCTGACGCCCGCCAGTGGTACGTGGACAGGCTGCGCGCTCTTCAAAAGTCTAATGGAATCGACAGTTTTAAATTCGATGGAGGAGAATCGAGTTGGTCTCCACAAATACCCGTCCTTCGAGGGGACATCAGAGACCAACCTGGTGTAATCACTGCCGACTACGTGAGGGCTGTGGCTGAGTTCGGCCCGATGGTAGAAGTCAGAGCTGGATACAGGACCCAAGACCTCCCGGTCTTCGTGCGAATGATAGACAAGGACAGCTACTGGACCTTCGAGAATGGTCTTCCGACCGTCGTGACCACTTTGCTGCAGATGAACATGAACGGTTACCCCCTAGTCTTACCTGACATGATCGGAGGAAACGGGTACAACGAACCACCGACTAAAGAGTTGTTCATCAGGTGGCTCCAAGCCACAACGTTTATGCCTAGCATGCAGTTCAGCTATGTACCCTGGGACTTCGATAATGAGACAATTGAAATTAGCCAGAAATACGTGGCGCTCCACGCGAAGCACGCGGTCGACATTGTGGCCGCCTGCAAGCAAGCTACCCTGGACGGTACTCCTGTAAACCCTCCGATATGGTGGATCGCGCCCAACGATACCACAGCCTATGATATTTGGGATGAATATCTCCTGGGTGAGGGGATCTTGGTGGCCCCGGTGTTAGAGCGGGGGGCGAGGTCGCGTGACGTGTACTTGCCCCCGGGGACGTGGTTAGCGCAGGGAGACCCCGGCCGGGTGTTCCCCGGAGGCGACTGGATAAGAAAATACCCGGCACCTTTAGACACCTTACCGTATTTCATTCGGGCTTAA
Protein
MDAKIFLLLLSTVVFVNAVDISLIDSTDVKLKLQDNPYGGLNVLLNRRGISEVISVIGRRTSGIKDIIQNGDVLIQFKNDTTVRISVEHIAGERKGHLLSFAWKTTKNTRLEDCANLGTDSWYGGPQQKRQYWPIERLVLPNYSYVTKEADNSAVAEPYWLNSQGIFYYFDKKVPLFIDQNDLEKNAACFIAQIRPPYSKNRESNELVYALGIFGDVKQAHQFAVEKYLNKPKAIPDERMIKYPIWSTWARYKRDVNHDVVLKFADEINKNGFPNSQMEIDDLWEICYGSQTVDIRRFPDMKDMVGKLKAKGFRVTLWTHPFINKGCEPWYSEAKAKGYLVSSEYGSVETSWWNDNGTTTSYIDFTKPDARQWYVDRLRALQKSNGIDSFKFDGGESSWSPQIPVLRGDIRDQPGVITADYVRAVAEFGPMVEVRAGYRTQDLPVFVRMIDKDSYWTFENGLPTVVTTLLQMNMNGYPLVLPDMIGGNGYNEPPTKELFIRWLQATTFMPSMQFSYVPWDFDNETIEISQKYVALHAKHAVDIVAACKQATLDGTPVNPPIWWIAPNDTTAYDIWDEYLLGEGILVAPVLERGARSRDVYLPPGTWLAQGDPGRVFPGGDWIRKYPAPLDTLPYFIRA
Summary
Similarity
Belongs to the glycosyl hydrolase 31 family.
Uniprot
H9JCF4
A0A2W1BQL7
A0A2A4JKL7
A0A2H1V7R0
A0A3S2NV62
A0A212F9Y1
+ More
A0A2W1BJ10 A0A194QHP5 H9JCA8 A0A212F9W1 A0A2A4JKP3 A0A3S2TGG7 A0A0N1IHR0 A0A2W1BP21 A0A212F9W4 A0A437B4A7 A0A2W1BJI2 U5EPD4 A0A2A4JLH7 A0A1L8DQS6 A0A1L8DQS4 A0A1Q3FS49 A0A1L8DQQ2 A0A1L8DR05 A0A1S4F7Q1 A0A034WC76 W8BW19 B0XBN3 A0A0K8U7T4 Q17D16 A0A0K8UJF6 A0A1B6HX93 A0A1B6M652 F4ZHB3 A0A182H6M3 A0A1S4F7D7 A0A1B6GJB3 A0A2P8XH20 A0A059WND1 A0A0L7L4C9 Q19P00 A0A1B0DC58 A0A084W5D2 A0A182M216 A0A182KB80 A0A0L0BNZ0 A0A182VXM7 A0A1I8MXX3 A0A182I891 A0A182JJV8 B4KBX6 A0A1J1IRK5 A0A336KI79 A0A1Y1L5J4 A0A182XH62 A0A1I8NZK2 B4LX60 A0A182NNA4 A0A182QD29 A0A182KMP9 A0A1I8QDH1 A0A1I8JUQ8 A0A336LUM5 Q7PJP4 A0A0J7KND3 A0A336MDN0 T1PDF4 A0A182UMR5 A0A182YIE3 A0A182PF15 A0A182TWE5 A0A1I8PKN7 A0A182F830 E2BIF0 B4NIJ4 A0A067QSX5 A0A026W2R2 A0A3L8DWT2 B4JFN1 A0A1B6CDQ5 A0A151J976 U5EUS8 A0A182YIE7 B4QUV6 B3LW43 A0A182MV42 A0A1S4GKB0 A0A084W5C4 Q7Q275 A0A195FCL9 A0A0N1IDN6 A0A182KMQ1 A0A182XH57 A0A182VJL5 A0A182F833 A0A182TB72 A0A182Q7J8 A0A182TPL7 A0A182I895 A0A1Q3FLK5
A0A2W1BJ10 A0A194QHP5 H9JCA8 A0A212F9W1 A0A2A4JKP3 A0A3S2TGG7 A0A0N1IHR0 A0A2W1BP21 A0A212F9W4 A0A437B4A7 A0A2W1BJI2 U5EPD4 A0A2A4JLH7 A0A1L8DQS6 A0A1L8DQS4 A0A1Q3FS49 A0A1L8DQQ2 A0A1L8DR05 A0A1S4F7Q1 A0A034WC76 W8BW19 B0XBN3 A0A0K8U7T4 Q17D16 A0A0K8UJF6 A0A1B6HX93 A0A1B6M652 F4ZHB3 A0A182H6M3 A0A1S4F7D7 A0A1B6GJB3 A0A2P8XH20 A0A059WND1 A0A0L7L4C9 Q19P00 A0A1B0DC58 A0A084W5D2 A0A182M216 A0A182KB80 A0A0L0BNZ0 A0A182VXM7 A0A1I8MXX3 A0A182I891 A0A182JJV8 B4KBX6 A0A1J1IRK5 A0A336KI79 A0A1Y1L5J4 A0A182XH62 A0A1I8NZK2 B4LX60 A0A182NNA4 A0A182QD29 A0A182KMP9 A0A1I8QDH1 A0A1I8JUQ8 A0A336LUM5 Q7PJP4 A0A0J7KND3 A0A336MDN0 T1PDF4 A0A182UMR5 A0A182YIE3 A0A182PF15 A0A182TWE5 A0A1I8PKN7 A0A182F830 E2BIF0 B4NIJ4 A0A067QSX5 A0A026W2R2 A0A3L8DWT2 B4JFN1 A0A1B6CDQ5 A0A151J976 U5EUS8 A0A182YIE7 B4QUV6 B3LW43 A0A182MV42 A0A1S4GKB0 A0A084W5C4 Q7Q275 A0A195FCL9 A0A0N1IDN6 A0A182KMQ1 A0A182XH57 A0A182VJL5 A0A182F833 A0A182TB72 A0A182Q7J8 A0A182TPL7 A0A182I895 A0A1Q3FLK5
Pubmed
EMBL
BABH01031586
KZ150013
PZC75030.1
NWSH01001097
PCG72625.1
ODYU01001088
+ More
SOQ36836.1 RSAL01000164 RVE45336.1 AGBW02009556 OWR50519.1 PZC75029.1 KQ458981 KPJ04460.1 BABH01031610 BABH01031611 OWR50521.1 PCG72627.1 RVE45338.1 LADJ01009642 KPJ20937.1 PZC75027.1 OWR50520.1 RVE45337.1 PZC75028.1 GANO01003733 JAB56138.1 PCG72626.1 GFDF01005272 JAV08812.1 GFDF01005271 JAV08813.1 GFDL01004729 JAV30316.1 GFDF01005301 JAV08783.1 GFDF01005277 JAV08807.1 GAKP01007549 JAC51403.1 GAMC01013151 JAB93404.1 DS232644 EDS44382.1 GDHF01029666 JAI22648.1 CH477301 EAT44243.1 GDHF01025510 JAI26804.1 GECU01028408 JAS79298.1 GEBQ01008596 JAT31381.1 HQ015443 ADL40394.1 JXUM01114446 KQ565787 KXJ70664.1 GECZ01007254 JAS62515.1 PYGN01002153 PSN31308.1 KJ576837 AIA09350.1 JTDY01003042 KOB70265.1 DQ515931 ABF71570.1 AJVK01030572 AJVK01030573 AJVK01030574 ATLV01020585 KE525303 KFB45426.1 AXCM01014517 JRES01001582 KNC21726.1 APCN01002678 APCN01002679 CH933806 EDW15828.1 CVRI01000058 CRL02784.1 UFQS01000448 UFQT01000448 SSX04071.1 SSX24436.1 GEZM01063873 JAV68942.1 CH940650 EDW66712.1 AXCN02001904 UFQS01000118 UFQT01000118 SSW99999.1 SSX20379.1 AAAB01008978 EAA43680.4 LBMM01005028 KMQ91888.1 UFQT01000758 SSX27019.1 KA646719 AFP61348.1 GL448504 EFN84538.1 CH964272 EDW84817.1 KK853055 KDR11965.1 KK107459 EZA50367.1 QOIP01000004 RLU24248.1 CH916369 EDV93512.1 GEDC01025706 JAS11592.1 KQ979433 KYN21584.1 GANO01003734 JAB56137.1 CM000364 EDX14422.1 CH902617 EDV42621.1 AXCM01009062 ATLV01020576 ATLV01020577 ATLV01020578 KFB45418.1 EAA13629.5 KQ981673 KYN38385.1 LADJ01000497 KPJ20835.1 AXCN02000286 APCN01002681 APCN01002682 GFDL01006647 JAV28398.1
SOQ36836.1 RSAL01000164 RVE45336.1 AGBW02009556 OWR50519.1 PZC75029.1 KQ458981 KPJ04460.1 BABH01031610 BABH01031611 OWR50521.1 PCG72627.1 RVE45338.1 LADJ01009642 KPJ20937.1 PZC75027.1 OWR50520.1 RVE45337.1 PZC75028.1 GANO01003733 JAB56138.1 PCG72626.1 GFDF01005272 JAV08812.1 GFDF01005271 JAV08813.1 GFDL01004729 JAV30316.1 GFDF01005301 JAV08783.1 GFDF01005277 JAV08807.1 GAKP01007549 JAC51403.1 GAMC01013151 JAB93404.1 DS232644 EDS44382.1 GDHF01029666 JAI22648.1 CH477301 EAT44243.1 GDHF01025510 JAI26804.1 GECU01028408 JAS79298.1 GEBQ01008596 JAT31381.1 HQ015443 ADL40394.1 JXUM01114446 KQ565787 KXJ70664.1 GECZ01007254 JAS62515.1 PYGN01002153 PSN31308.1 KJ576837 AIA09350.1 JTDY01003042 KOB70265.1 DQ515931 ABF71570.1 AJVK01030572 AJVK01030573 AJVK01030574 ATLV01020585 KE525303 KFB45426.1 AXCM01014517 JRES01001582 KNC21726.1 APCN01002678 APCN01002679 CH933806 EDW15828.1 CVRI01000058 CRL02784.1 UFQS01000448 UFQT01000448 SSX04071.1 SSX24436.1 GEZM01063873 JAV68942.1 CH940650 EDW66712.1 AXCN02001904 UFQS01000118 UFQT01000118 SSW99999.1 SSX20379.1 AAAB01008978 EAA43680.4 LBMM01005028 KMQ91888.1 UFQT01000758 SSX27019.1 KA646719 AFP61348.1 GL448504 EFN84538.1 CH964272 EDW84817.1 KK853055 KDR11965.1 KK107459 EZA50367.1 QOIP01000004 RLU24248.1 CH916369 EDV93512.1 GEDC01025706 JAS11592.1 KQ979433 KYN21584.1 GANO01003734 JAB56137.1 CM000364 EDX14422.1 CH902617 EDV42621.1 AXCM01009062 ATLV01020576 ATLV01020577 ATLV01020578 KFB45418.1 EAA13629.5 KQ981673 KYN38385.1 LADJ01000497 KPJ20835.1 AXCN02000286 APCN01002681 APCN01002682 GFDL01006647 JAV28398.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
+ More
UP000002320 UP000008820 UP000069940 UP000249989 UP000245037 UP000037510 UP000092462 UP000030765 UP000075883 UP000075881 UP000037069 UP000075920 UP000095301 UP000075840 UP000075880 UP000009192 UP000183832 UP000076407 UP000095300 UP000008792 UP000075884 UP000075886 UP000075882 UP000075900 UP000007062 UP000036403 UP000075903 UP000076408 UP000075885 UP000075902 UP000069272 UP000008237 UP000007798 UP000027135 UP000053097 UP000279307 UP000001070 UP000078492 UP000000304 UP000007801 UP000078541 UP000075901
UP000002320 UP000008820 UP000069940 UP000249989 UP000245037 UP000037510 UP000092462 UP000030765 UP000075883 UP000075881 UP000037069 UP000075920 UP000095301 UP000075840 UP000075880 UP000009192 UP000183832 UP000076407 UP000095300 UP000008792 UP000075884 UP000075886 UP000075882 UP000075900 UP000007062 UP000036403 UP000075903 UP000076408 UP000075885 UP000075902 UP000069272 UP000008237 UP000007798 UP000027135 UP000053097 UP000279307 UP000001070 UP000078492 UP000000304 UP000007801 UP000078541 UP000075901
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
H9JCF4
A0A2W1BQL7
A0A2A4JKL7
A0A2H1V7R0
A0A3S2NV62
A0A212F9Y1
+ More
A0A2W1BJ10 A0A194QHP5 H9JCA8 A0A212F9W1 A0A2A4JKP3 A0A3S2TGG7 A0A0N1IHR0 A0A2W1BP21 A0A212F9W4 A0A437B4A7 A0A2W1BJI2 U5EPD4 A0A2A4JLH7 A0A1L8DQS6 A0A1L8DQS4 A0A1Q3FS49 A0A1L8DQQ2 A0A1L8DR05 A0A1S4F7Q1 A0A034WC76 W8BW19 B0XBN3 A0A0K8U7T4 Q17D16 A0A0K8UJF6 A0A1B6HX93 A0A1B6M652 F4ZHB3 A0A182H6M3 A0A1S4F7D7 A0A1B6GJB3 A0A2P8XH20 A0A059WND1 A0A0L7L4C9 Q19P00 A0A1B0DC58 A0A084W5D2 A0A182M216 A0A182KB80 A0A0L0BNZ0 A0A182VXM7 A0A1I8MXX3 A0A182I891 A0A182JJV8 B4KBX6 A0A1J1IRK5 A0A336KI79 A0A1Y1L5J4 A0A182XH62 A0A1I8NZK2 B4LX60 A0A182NNA4 A0A182QD29 A0A182KMP9 A0A1I8QDH1 A0A1I8JUQ8 A0A336LUM5 Q7PJP4 A0A0J7KND3 A0A336MDN0 T1PDF4 A0A182UMR5 A0A182YIE3 A0A182PF15 A0A182TWE5 A0A1I8PKN7 A0A182F830 E2BIF0 B4NIJ4 A0A067QSX5 A0A026W2R2 A0A3L8DWT2 B4JFN1 A0A1B6CDQ5 A0A151J976 U5EUS8 A0A182YIE7 B4QUV6 B3LW43 A0A182MV42 A0A1S4GKB0 A0A084W5C4 Q7Q275 A0A195FCL9 A0A0N1IDN6 A0A182KMQ1 A0A182XH57 A0A182VJL5 A0A182F833 A0A182TB72 A0A182Q7J8 A0A182TPL7 A0A182I895 A0A1Q3FLK5
A0A2W1BJ10 A0A194QHP5 H9JCA8 A0A212F9W1 A0A2A4JKP3 A0A3S2TGG7 A0A0N1IHR0 A0A2W1BP21 A0A212F9W4 A0A437B4A7 A0A2W1BJI2 U5EPD4 A0A2A4JLH7 A0A1L8DQS6 A0A1L8DQS4 A0A1Q3FS49 A0A1L8DQQ2 A0A1L8DR05 A0A1S4F7Q1 A0A034WC76 W8BW19 B0XBN3 A0A0K8U7T4 Q17D16 A0A0K8UJF6 A0A1B6HX93 A0A1B6M652 F4ZHB3 A0A182H6M3 A0A1S4F7D7 A0A1B6GJB3 A0A2P8XH20 A0A059WND1 A0A0L7L4C9 Q19P00 A0A1B0DC58 A0A084W5D2 A0A182M216 A0A182KB80 A0A0L0BNZ0 A0A182VXM7 A0A1I8MXX3 A0A182I891 A0A182JJV8 B4KBX6 A0A1J1IRK5 A0A336KI79 A0A1Y1L5J4 A0A182XH62 A0A1I8NZK2 B4LX60 A0A182NNA4 A0A182QD29 A0A182KMP9 A0A1I8QDH1 A0A1I8JUQ8 A0A336LUM5 Q7PJP4 A0A0J7KND3 A0A336MDN0 T1PDF4 A0A182UMR5 A0A182YIE3 A0A182PF15 A0A182TWE5 A0A1I8PKN7 A0A182F830 E2BIF0 B4NIJ4 A0A067QSX5 A0A026W2R2 A0A3L8DWT2 B4JFN1 A0A1B6CDQ5 A0A151J976 U5EUS8 A0A182YIE7 B4QUV6 B3LW43 A0A182MV42 A0A1S4GKB0 A0A084W5C4 Q7Q275 A0A195FCL9 A0A0N1IDN6 A0A182KMQ1 A0A182XH57 A0A182VJL5 A0A182F833 A0A182TB72 A0A182Q7J8 A0A182TPL7 A0A182I895 A0A1Q3FLK5
PDB
6JR7
E-value=4.16265e-24,
Score=278
Ontologies
GO
Topology
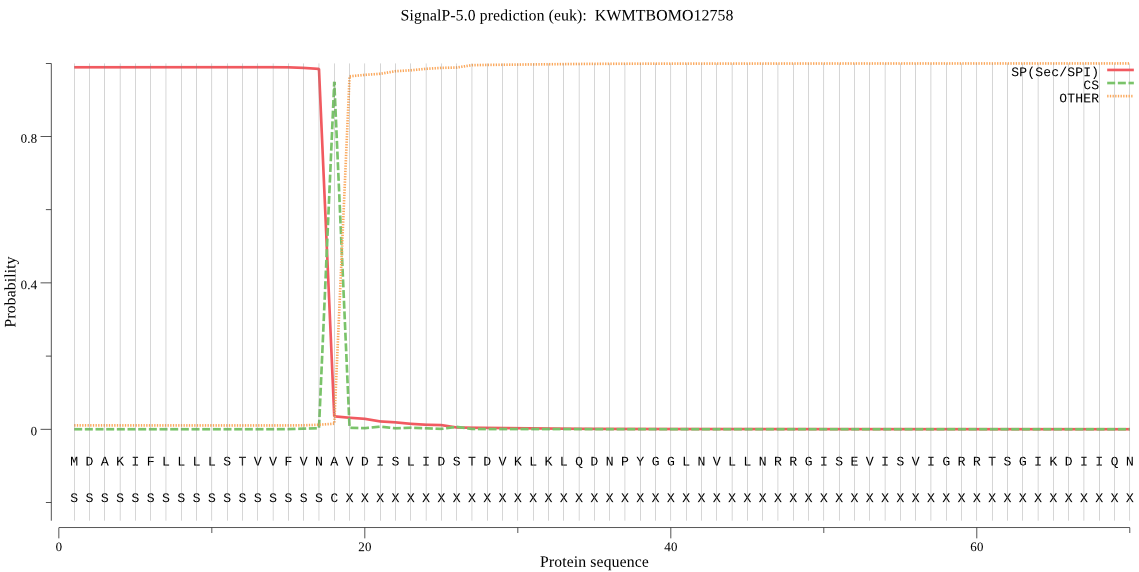
SignalP
Position: 1 - 18,
Likelihood: 0.988878
Length:
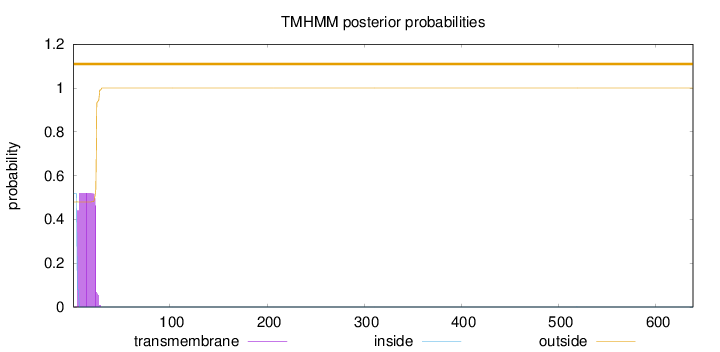
638
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.36773
Exp number, first 60 AAs:
10.36528
Total prob of N-in:
0.51995
POSSIBLE N-term signal
sequence
outside
1 - 638
Population Genetic Test Statistics
Pi
18.674189
Theta
18.907618
Tajima's D
-1.31254
CLR
0.721438
CSRT
0.0854957252137393
Interpretation
Uncertain
