Pre Gene Modal
BGIBMGA007195
Annotation
PREDICTED:_cytochrome_P450_4C1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.751 Nuclear Reliability : 1.117
Sequence
CDS
ATGATTTGGACTGTAGTTGTATTGATTATTTTTATATTCTTGATATGGAAGGCTTTAGAGGATGAGGAGAATCCTCTTGACTCTCTTCCCGGACCAGAAAGGAAGCCGATCATTGGAGCCGCATTGCGATTCGTCAATTTAAATACAAGAGAAATGTTTATAAAATTGCGCGAGTACCATGCGATGTACGGTAATAGATTCGTTGTAAAGATATTCAAGAGGAGAGTCCTGCATCTGTCTAACGAAAAGGATGCTGAGGTCGTATTGTCGCATTCGAAGAACATTAAGAAAAGCAAGGGCTATACGTTTCTGAGTCCTTGGCTCGGCAGCGGTCTCCTGTTGAGCACCGGTTTTAAATGGCACAGCAGACGTAAGATTCTGACACCAACGTTTCACTTTAACATCTTGAAGAGCTTCTTAGAAATCATCAAGGACAAGAGTTGTGACTTGGTGAAGAGATTGGAGGAGTACAGGGGCGAAGAAGTAGACTTGATGCCCGTTATCAGCGACTTTACATTGTATACGATTTGCGAAACTGCAATGGGCACCCAATTGGATTCAGACAAGTCAGCAGAAACCAAGGAATACAAAATGGCCATACTACAAATAGCATCCTTGCTGCTCAACAGACTGACCAAAGTTTGGCTCCACAACGACTTAATATTCGACCAGCTTACAGTTGGGCGAAAGTTTCAGAAGTGCCTCAAACAAGTCCACTCTTTCGCCCATAATGTTATAGTTGAGAGGAAACGACAACGAGCATCTGGTAGAGACTATTCTGTCGTTGCAGAAGATGTATTCGGTCGTAAAAAGAGATTGGCCATGTTAGATCTTTTATTGGAAGCGGAAGAGAAAAATGAAATTGATTTTGAAGGAATAATGGATGAAGTTAATACATTTATGTTTGAGGGACACGACACAACTGCAGTCGCTTTAACGTTCAGTTTGATGTTGGTCGCGGAGGACGATCAAGTTCAAGATCGCATTTACAAGGAACTGCAAGGGATTTTTGGAGACTCTGACCGTCGGCCAACAATCAGTGATGTAGCCGAGATGAAGTACTTGGAAGCTGTAGTGAAGGAGACTCTGAGGCTGTATCCAAGTGTCCCGTTCATCGCTCGAGAGATCACTGAGGACTTTATGCTAGATGACCTAAACATAAAGAAAGGCTCAGAAGTAGTAGTCCACATATATGATCTGCATCGTCGTAAAGAGCTGTTTGCTGACCCCGAGAAGTTCCAGCCCGATAGATTCTTGAACGGAGAACTGAAGCATCCATACAGCTTCGTGCCCTTTAGCGCAGGACCAAGGAACTGTATTGGTCAACGCTTCGCGATGCTGGAAATGAAGTGCGTCCTCAGTGAGATCTGTCGTAGCTTCAGACTAGAACCCCGCACTAAGGGTTGGCGACCTACCCTCGTAGCGGAAATGCTTTTACGACCAAACGAACCTATTCATGTAAAGTTTATTAAACGCAAACAAAGCTGA
Protein
MIWTVVVLIIFIFLIWKALEDEENPLDSLPGPERKPIIGAALRFVNLNTREMFIKLREYHAMYGNRFVVKIFKRRVLHLSNEKDAEVVLSHSKNIKKSKGYTFLSPWLGSGLLLSTGFKWHSRRKILTPTFHFNILKSFLEIIKDKSCDLVKRLEEYRGEEVDLMPVISDFTLYTICETAMGTQLDSDKSAETKEYKMAILQIASLLLNRLTKVWLHNDLIFDQLTVGRKFQKCLKQVHSFAHNVIVERKRQRASGRDYSVVAEDVFGRKKRLAMLDLLLEAEEKNEIDFEGIMDEVNTFMFEGHDTTAVALTFSLMLVAEDDQVQDRIYKELQGIFGDSDRRPTISDVAEMKYLEAVVKETLRLYPSVPFIAREITEDFMLDDLNIKKGSEVVVHIYDLHRRKELFADPEKFQPDRFLNGELKHPYSFVPFSAGPRNCIGQRFAMLEMKCVLSEICRSFRLEPRTKGWRPTLVAEMLLRPNEPIHVKFIKRKQS
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
L0N6M8
L0N716
Q8ISJ8
A0A2W1BLR4
B2BNZ3
Q2I0J0
+ More
A0A248QHK4 J7FIT5 A0A286QUL0 A0A1E1W5V0 A0A194QLZ7 A0A0N1IEQ4 A0A068EU59 A0A0U2DUD7 A0A068EVK2 I4DP28 A0A2A4ITL8 A0A1V0D9C7 A0A0N0PBM3 H9JWX8 L0N7C1 H9JCC3 A0A2H1WM10 A0A1E1WH40 A0A0C5C1N3 D5L0M0 C1LZ55 L0N4I2 L0N5E9 A0A248QJ75 A0A248QFF8 A5A3F8 A5HG32 Q8MTC9 A0A2H1V2B5 A9QW14 D0FZ08 A0A2W1BJD8 L0N790 A0A2A4JUK6 A0A212ET62 Q2L7F2 A0MNW3 A0A1E1WM70 W5JEA0 A0A182G0G5 A0A068ETT9 A0A437B7J2 D5L0M2 X5DQR3 Q25508 A0A068F0W3 A0A172MA26 A0A2W1BPC2 X5DT35 A0MNW4 A0A212F1V8 A0A437B7Q4 A0A182S536 D5L0M1 A0A423TV42 A0A3S2NFU8 A0A286MXM1 A0A2A4JU89 V5W638 A0A1E1WBF9 A0A0K0LBB5 A0A212EXA9 B3GVW3 A0A411E526 A0A0K0LBB1 U5EWR0 Q6VT81 A0A0P4W0N6 D4PEE6 A0A0P4W5K9 A0A182PZ05 U5EWH6 A0A288ZBI1 A0A182G0G4 A0A182LYA3 A0A0P4W8L7 A0A1U8YID0 A0A248QEJ1 K4FC60 A0A194QYS2 A0A1Q1NKY5 A0A0K0LBC0 A0A0K0LBA8 A0A0C5BXG0 A0A2Y9D1T1 A0A084WPX4 Q7PIH4 Q8MTD0 A0A182XUC6 A0A182J268 A0A182VPL2
A0A248QHK4 J7FIT5 A0A286QUL0 A0A1E1W5V0 A0A194QLZ7 A0A0N1IEQ4 A0A068EU59 A0A0U2DUD7 A0A068EVK2 I4DP28 A0A2A4ITL8 A0A1V0D9C7 A0A0N0PBM3 H9JWX8 L0N7C1 H9JCC3 A0A2H1WM10 A0A1E1WH40 A0A0C5C1N3 D5L0M0 C1LZ55 L0N4I2 L0N5E9 A0A248QJ75 A0A248QFF8 A5A3F8 A5HG32 Q8MTC9 A0A2H1V2B5 A9QW14 D0FZ08 A0A2W1BJD8 L0N790 A0A2A4JUK6 A0A212ET62 Q2L7F2 A0MNW3 A0A1E1WM70 W5JEA0 A0A182G0G5 A0A068ETT9 A0A437B7J2 D5L0M2 X5DQR3 Q25508 A0A068F0W3 A0A172MA26 A0A2W1BPC2 X5DT35 A0MNW4 A0A212F1V8 A0A437B7Q4 A0A182S536 D5L0M1 A0A423TV42 A0A3S2NFU8 A0A286MXM1 A0A2A4JU89 V5W638 A0A1E1WBF9 A0A0K0LBB5 A0A212EXA9 B3GVW3 A0A411E526 A0A0K0LBB1 U5EWR0 Q6VT81 A0A0P4W0N6 D4PEE6 A0A0P4W5K9 A0A182PZ05 U5EWH6 A0A288ZBI1 A0A182G0G4 A0A182LYA3 A0A0P4W8L7 A0A1U8YID0 A0A248QEJ1 K4FC60 A0A194QYS2 A0A1Q1NKY5 A0A0K0LBC0 A0A0K0LBA8 A0A0C5BXG0 A0A2Y9D1T1 A0A084WPX4 Q7PIH4 Q8MTD0 A0A182XUC6 A0A182J268 A0A182VPL2
Pubmed
EMBL
AK343220
BAM73902.1
AK289284
BAM73811.1
AY063500
AAL48299.1
+ More
KZ150005 PZC75221.1 EF591060 ABU88427.1 DQ355383 ABC84370.2 KX443462 ASO98037.1 JX310089 AFP20600.1 KX443461 ASO98036.1 GDQN01008735 GDQN01003028 GDQN01001407 JAT82319.1 JAT88026.1 JAT89647.1 KQ458981 KPJ04476.1 KQ460882 KPJ11479.1 KM016732 AID54884.1 PZC75222.1 KP058451 AKP80586.1 KM016731 AID54883.1 AK403264 BAM19668.1 NWSH01006615 PCG63327.1 KY212039 ARA91603.1 KPJ11478.1 BABH01041553 AK343191 BAM73880.1 BABH01031558 BABH01031559 BABH01031560 ODYU01009575 SOQ54099.1 GDQN01004730 GDQN01002632 GDQN01000042 JAT86324.1 JAT88422.1 JAT91012.1 KP001139 AJN91184.1 GU731525 ADE05575.1 FN356972 CAX94850.1 AK289282 BAM73809.1 AK289281 BAM73808.1 KX443466 ASO98041.1 KX443467 ASO98042.1 EF523434 ABP87671.1 EF528484 ABP99018.1 AY113688 AAM54723.1 ODYU01000355 SOQ34985.1 SOQ34986.1 EU273876 ABX64439.1 AB498590 BAI45222.1 KZ150007 PZC75159.1 AK289283 BAM73810.1 NWSH01000604 PCG75378.1 AGBW02012640 OWR44670.1 DQ352137 ABC72321.2 EF017796 ABK27871.1 GDQN01009628 GDQN01002954 JAT81426.1 JAT88100.1 ADMH02001390 ETN62682.1 KM016730 AID54882.1 RSAL01000129 RVE46480.1 GU731527 ADE05577.1 KF701163 AHW57333.1 L38671 AAC21661.1 KM016728 AID54880.1 KX008607 ANC90156.1 PZC75157.1 KF701164 AHW57334.1 EF017797 ABK27872.1 AGBW02010786 OWR47720.1 RVE46478.1 GU731526 ADE05576.1 QCYY01001130 ROT80325.1 RVE46477.1 MF684339 ASX93973.1 PCG75379.1 KF668286 AHC02483.1 GDQN01006741 JAT84313.1 KM217034 AIW79997.1 AGBW02011790 OWR46123.1 FM163384 CAQ57674.1 MH236443 QBA57448.1 KM217037 AIW80000.1 GANO01001299 JAB58572.1 AY328467 AAQ93010.1 GDRN01081744 JAI61950.1 GU969106 ADD63783.1 GDRN01081739 JAI61953.1 GANO01002947 JAB56924.1 KX812713 ASU43935.1 AXCM01004515 GDRN01081741 JAI61951.1 KR057738 AMP19274.1 KX443468 ASO98043.1 JN835470 AFH35031.1 KQ461150 KPJ08736.1 KX660697 AQM57036.1 KM217039 AIW80002.1 KM217038 AIW80001.1 KP001143 AJN91188.1 ATLV01025136 KE525369 KFB52268.1 AAAB01008960 EAA44145.4 AY113687 AAM54722.1
KZ150005 PZC75221.1 EF591060 ABU88427.1 DQ355383 ABC84370.2 KX443462 ASO98037.1 JX310089 AFP20600.1 KX443461 ASO98036.1 GDQN01008735 GDQN01003028 GDQN01001407 JAT82319.1 JAT88026.1 JAT89647.1 KQ458981 KPJ04476.1 KQ460882 KPJ11479.1 KM016732 AID54884.1 PZC75222.1 KP058451 AKP80586.1 KM016731 AID54883.1 AK403264 BAM19668.1 NWSH01006615 PCG63327.1 KY212039 ARA91603.1 KPJ11478.1 BABH01041553 AK343191 BAM73880.1 BABH01031558 BABH01031559 BABH01031560 ODYU01009575 SOQ54099.1 GDQN01004730 GDQN01002632 GDQN01000042 JAT86324.1 JAT88422.1 JAT91012.1 KP001139 AJN91184.1 GU731525 ADE05575.1 FN356972 CAX94850.1 AK289282 BAM73809.1 AK289281 BAM73808.1 KX443466 ASO98041.1 KX443467 ASO98042.1 EF523434 ABP87671.1 EF528484 ABP99018.1 AY113688 AAM54723.1 ODYU01000355 SOQ34985.1 SOQ34986.1 EU273876 ABX64439.1 AB498590 BAI45222.1 KZ150007 PZC75159.1 AK289283 BAM73810.1 NWSH01000604 PCG75378.1 AGBW02012640 OWR44670.1 DQ352137 ABC72321.2 EF017796 ABK27871.1 GDQN01009628 GDQN01002954 JAT81426.1 JAT88100.1 ADMH02001390 ETN62682.1 KM016730 AID54882.1 RSAL01000129 RVE46480.1 GU731527 ADE05577.1 KF701163 AHW57333.1 L38671 AAC21661.1 KM016728 AID54880.1 KX008607 ANC90156.1 PZC75157.1 KF701164 AHW57334.1 EF017797 ABK27872.1 AGBW02010786 OWR47720.1 RVE46478.1 GU731526 ADE05576.1 QCYY01001130 ROT80325.1 RVE46477.1 MF684339 ASX93973.1 PCG75379.1 KF668286 AHC02483.1 GDQN01006741 JAT84313.1 KM217034 AIW79997.1 AGBW02011790 OWR46123.1 FM163384 CAQ57674.1 MH236443 QBA57448.1 KM217037 AIW80000.1 GANO01001299 JAB58572.1 AY328467 AAQ93010.1 GDRN01081744 JAI61950.1 GU969106 ADD63783.1 GDRN01081739 JAI61953.1 GANO01002947 JAB56924.1 KX812713 ASU43935.1 AXCM01004515 GDRN01081741 JAI61951.1 KR057738 AMP19274.1 KX443468 ASO98043.1 JN835470 AFH35031.1 KQ461150 KPJ08736.1 KX660697 AQM57036.1 KM217039 AIW80002.1 KM217038 AIW80001.1 KP001143 AJN91188.1 ATLV01025136 KE525369 KFB52268.1 AAAB01008960 EAA44145.4 AY113687 AAM54722.1
Proteomes
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
L0N6M8
L0N716
Q8ISJ8
A0A2W1BLR4
B2BNZ3
Q2I0J0
+ More
A0A248QHK4 J7FIT5 A0A286QUL0 A0A1E1W5V0 A0A194QLZ7 A0A0N1IEQ4 A0A068EU59 A0A0U2DUD7 A0A068EVK2 I4DP28 A0A2A4ITL8 A0A1V0D9C7 A0A0N0PBM3 H9JWX8 L0N7C1 H9JCC3 A0A2H1WM10 A0A1E1WH40 A0A0C5C1N3 D5L0M0 C1LZ55 L0N4I2 L0N5E9 A0A248QJ75 A0A248QFF8 A5A3F8 A5HG32 Q8MTC9 A0A2H1V2B5 A9QW14 D0FZ08 A0A2W1BJD8 L0N790 A0A2A4JUK6 A0A212ET62 Q2L7F2 A0MNW3 A0A1E1WM70 W5JEA0 A0A182G0G5 A0A068ETT9 A0A437B7J2 D5L0M2 X5DQR3 Q25508 A0A068F0W3 A0A172MA26 A0A2W1BPC2 X5DT35 A0MNW4 A0A212F1V8 A0A437B7Q4 A0A182S536 D5L0M1 A0A423TV42 A0A3S2NFU8 A0A286MXM1 A0A2A4JU89 V5W638 A0A1E1WBF9 A0A0K0LBB5 A0A212EXA9 B3GVW3 A0A411E526 A0A0K0LBB1 U5EWR0 Q6VT81 A0A0P4W0N6 D4PEE6 A0A0P4W5K9 A0A182PZ05 U5EWH6 A0A288ZBI1 A0A182G0G4 A0A182LYA3 A0A0P4W8L7 A0A1U8YID0 A0A248QEJ1 K4FC60 A0A194QYS2 A0A1Q1NKY5 A0A0K0LBC0 A0A0K0LBA8 A0A0C5BXG0 A0A2Y9D1T1 A0A084WPX4 Q7PIH4 Q8MTD0 A0A182XUC6 A0A182J268 A0A182VPL2
A0A248QHK4 J7FIT5 A0A286QUL0 A0A1E1W5V0 A0A194QLZ7 A0A0N1IEQ4 A0A068EU59 A0A0U2DUD7 A0A068EVK2 I4DP28 A0A2A4ITL8 A0A1V0D9C7 A0A0N0PBM3 H9JWX8 L0N7C1 H9JCC3 A0A2H1WM10 A0A1E1WH40 A0A0C5C1N3 D5L0M0 C1LZ55 L0N4I2 L0N5E9 A0A248QJ75 A0A248QFF8 A5A3F8 A5HG32 Q8MTC9 A0A2H1V2B5 A9QW14 D0FZ08 A0A2W1BJD8 L0N790 A0A2A4JUK6 A0A212ET62 Q2L7F2 A0MNW3 A0A1E1WM70 W5JEA0 A0A182G0G5 A0A068ETT9 A0A437B7J2 D5L0M2 X5DQR3 Q25508 A0A068F0W3 A0A172MA26 A0A2W1BPC2 X5DT35 A0MNW4 A0A212F1V8 A0A437B7Q4 A0A182S536 D5L0M1 A0A423TV42 A0A3S2NFU8 A0A286MXM1 A0A2A4JU89 V5W638 A0A1E1WBF9 A0A0K0LBB5 A0A212EXA9 B3GVW3 A0A411E526 A0A0K0LBB1 U5EWR0 Q6VT81 A0A0P4W0N6 D4PEE6 A0A0P4W5K9 A0A182PZ05 U5EWH6 A0A288ZBI1 A0A182G0G4 A0A182LYA3 A0A0P4W8L7 A0A1U8YID0 A0A248QEJ1 K4FC60 A0A194QYS2 A0A1Q1NKY5 A0A0K0LBC0 A0A0K0LBA8 A0A0C5BXG0 A0A2Y9D1T1 A0A084WPX4 Q7PIH4 Q8MTD0 A0A182XUC6 A0A182J268 A0A182VPL2
PDB
6C94
E-value=5.18249e-50,
Score=500
Ontologies
GO
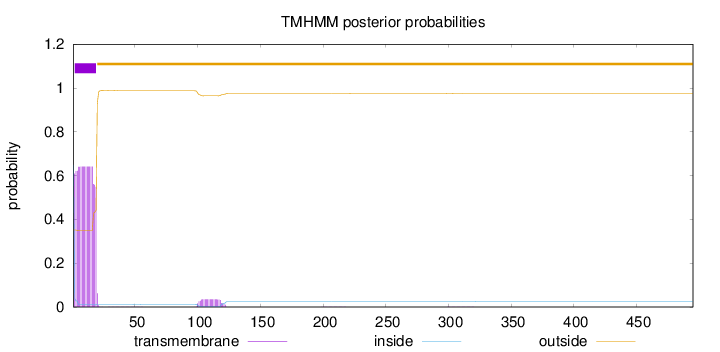
Topology
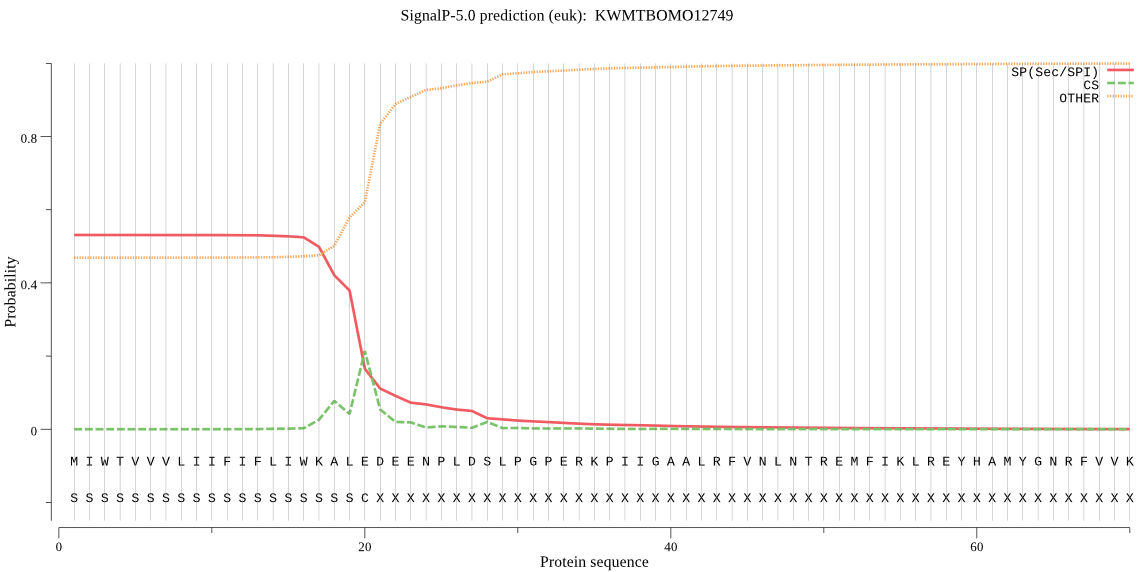
SignalP
Position: 1 - 20,
Likelihood: 0.531075
Length:
495
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
11.9895899999999
Exp number, first 60 AAs:
11.29533
Total prob of N-in:
0.64880
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 19
outside
20 - 495
Population Genetic Test Statistics
Pi
3.144465
Theta
3.439262
Tajima's D
-1.128038
CLR
1.290388
CSRT
0.112844357782111
Interpretation
Uncertain
