Gene
KWMTBOMO12745
Annotation
integrase_core_domain_protein_[Lasius_niger]
Location in the cell
Nuclear Reliability : 3.372
Sequence
CDS
ATGAAAAATGCCAAATCTTGTGACACGCCCGCAAGTAGCGATTCAAAGTCGATACAGGAAACTGCCCAGGAAAAGGAATATCCCCATAGGGAAGCACTGGGAAGTTTACTGTACCTGTCTACCAGAACCAGACCAGATATCGCACAAGCGGTCAATCTAGTTAGCAGGAAGGTAGAAAACTCCACTAGAGAAGACGTCACAAAAGTCAAGAAAATATTCAAATATTTAGTGGGAACCAAAGAGAAAGGCATTTTGTTCAAACGAAATAAACCGATAGAGAATATCGATAGTTACAGAGACTCGGATGATGCTGGAGATCCAAAACGCAAAGAAGCACTTCTGGCTCCGTGCTAA
Protein
MKNAKSCDTPASSDSKSIQETAQEKEYPHREALGSLLYLSTRTRPDIAQAVNLVSRKVENSTREDVTKVKKIFKYLVGTKEKGILFKRNKPIENIDSYRDSDDAGDPKRKEALLAPC
Summary
Uniprot
A0A0A9YBR9
A0A0A9Z091
A0A0A9ZF63
A0A0A9WDS7
A0A0A9W6J9
A0A0J7KC73
+ More
A0A329RKZ4 A0A0A9WDA6 A0A0J7K689 A0A0A9X3Z4 A0A2P4X1D7 A0A1B6GCK5 A0A085MSK2 A0A1B6FNY3 A0A0A9YCF4 A0A1B6H4J4 A0A146M1T4 A0A1Y1JZ65 A0A146LHR9 V5GE66 H3H8R6 A0A3M6V7W3 A0A0J7MNS8 H3GB86 H3H8M0 H3G8C4 H3H5N2 A0A1B6KG29 A0A1Y1KNM9 A0A329RR24 A0A0A9W2F2 A0A146LRM9 H3GB40 A0A0J7JZ17 H3H9X3 A0A0J7N098 H3GB42 H3GAY4 W4F9V3 H3GBA8 A0A0V0G477 H3G6C9 A0A0N5E2R3 A0A2P4YCF8 H3GAW7 H3H8S0 B3Y003 A0A1Y1KC03 A0A1X7U3D2 H3GZJ4 A0A0J7K9V7 A0A085MTW9 A0A1X7STY0 X1WST7 A0A1X1QS81 H3GAW9 H3H1B3 H9JF18 A0A151RHK1 A0A1X7SRH6 H3H922 A0A0A9XUC7 H3HA29 A0A139WJX6 H3GAT9 H3GY81 A0A2P4YV10 H3H949 A0A0B1PJ57 A0A085MSN1 A0A1Y1K7G3 A0A151SEX3 H3GIF1 K7JAV0 H3HAB2 A0A151U0S6 A0A2N9GS71 A0A151S124 A0A1B8AAN0 A0A087HHB1 H3GB53 X1WMK8 A0A182HAB6 A0A1Y1LVN7 H3H5F5 H3H9T4 A0A2P4WYA4 A0A3Q7HLK4 A0A2N9GKY6 A0A2P4YBB4 A0A1Y1NM77 A0A0A9X817 H3H6A9
A0A329RKZ4 A0A0A9WDA6 A0A0J7K689 A0A0A9X3Z4 A0A2P4X1D7 A0A1B6GCK5 A0A085MSK2 A0A1B6FNY3 A0A0A9YCF4 A0A1B6H4J4 A0A146M1T4 A0A1Y1JZ65 A0A146LHR9 V5GE66 H3H8R6 A0A3M6V7W3 A0A0J7MNS8 H3GB86 H3H8M0 H3G8C4 H3H5N2 A0A1B6KG29 A0A1Y1KNM9 A0A329RR24 A0A0A9W2F2 A0A146LRM9 H3GB40 A0A0J7JZ17 H3H9X3 A0A0J7N098 H3GB42 H3GAY4 W4F9V3 H3GBA8 A0A0V0G477 H3G6C9 A0A0N5E2R3 A0A2P4YCF8 H3GAW7 H3H8S0 B3Y003 A0A1Y1KC03 A0A1X7U3D2 H3GZJ4 A0A0J7K9V7 A0A085MTW9 A0A1X7STY0 X1WST7 A0A1X1QS81 H3GAW9 H3H1B3 H9JF18 A0A151RHK1 A0A1X7SRH6 H3H922 A0A0A9XUC7 H3HA29 A0A139WJX6 H3GAT9 H3GY81 A0A2P4YV10 H3H949 A0A0B1PJ57 A0A085MSN1 A0A1Y1K7G3 A0A151SEX3 H3GIF1 K7JAV0 H3HAB2 A0A151U0S6 A0A2N9GS71 A0A151S124 A0A1B8AAN0 A0A087HHB1 H3GB53 X1WMK8 A0A182HAB6 A0A1Y1LVN7 H3H5F5 H3H9T4 A0A2P4WYA4 A0A3Q7HLK4 A0A2N9GKY6 A0A2P4YBB4 A0A1Y1NM77 A0A0A9X817 H3H6A9
Pubmed
EMBL
GBHO01013072
JAG30532.1
GBHO01005715
JAG37889.1
GBHO01003079
JAG40525.1
+ More
GBHO01040574 JAG03030.1 GBHO01040573 JAG03031.1 LBMM01009639 KMQ87973.1 MJFZ01000952 RAW24002.1 GBHO01037840 JAG05764.1 LBMM01012772 KMQ86003.1 GBHO01031779 JAG11825.1 NCKW01017192 POM59370.1 GECZ01009601 JAS60168.1 KL367687 KFD60198.1 GECZ01017843 JAS51926.1 GBHO01016409 JAG27195.1 GECZ01000150 JAS69619.1 GDHC01004875 JAQ13754.1 GEZM01099971 JAV53080.1 GDHC01010945 JAQ07684.1 GALX01006157 JAB62309.1 DS567555 QLLG01000626 QKXF01000006 RMX62449.1 RQM18873.1 LBMM01027002 KMQ82250.1 DS567745 DS567497 DS566001 DS566383 GEBQ01029584 JAT10393.1 GEZM01078128 GEZM01078127 JAV63002.1 MJFZ01000618 RAW26780.1 GBHO01041007 JAG02597.1 GDHC01009233 JAQ09396.1 DS567087 LBMM01020254 KMQ83327.1 DS568207 LBMM01012672 KMQ86060.1 DS567139 DS566289 KI913645 ETV63694.1 DS568535 GECL01003258 JAP02866.1 DS568262 NCKW01003798 POM75488.1 DS566231 DS567564 AB378753 BAG68376.1 GEZM01093140 JAV56327.1 DS566082 LBMM01011114 KMQ87021.1 KL367655 KFD60665.1 ABLF02066511 LWTN01000174 ORU94050.1 DS566242 DS566098 BABH01033959 BABH01033960 KQ483738 KYP42039.1 DS567717 GBHO01019280 GBHO01019277 GBHO01019272 GBHO01014987 JAG24324.1 JAG24327.1 JAG24332.1 JAG28617.1 DS568300 KQ971338 KYB28117.1 DS566148 DS566071 NCKW01000038 POM81641.1 DS567770 KN538522 KHJ39825.1 KL367684 KFD60227.1 GEZM01090252 JAV57314.1 KQ483414 KYP53356.1 DS566011 AAZX01006115 AAZX01021021 DS568418 CM003604 KYP72919.1 OIVN01002268 SPD02134.1 KQ483498 KYP48513.1 LYXU01000014 OBS17533.1 CM002870 KFK41513.1 DS567227 ABLF02066512 JXUM01122536 KQ566615 KXJ69965.1 GEZM01048830 JAV76370.1 DS566328 DS568148 NCKW01020291 POM58281.1 OIVN01002376 SPD03107.1 NCKW01004050 POM75106.1 GEZM01002084 JAV97376.1 GBHO01028636 GBHO01028635 JAG14968.1 JAG14969.1 DS566528
GBHO01040574 JAG03030.1 GBHO01040573 JAG03031.1 LBMM01009639 KMQ87973.1 MJFZ01000952 RAW24002.1 GBHO01037840 JAG05764.1 LBMM01012772 KMQ86003.1 GBHO01031779 JAG11825.1 NCKW01017192 POM59370.1 GECZ01009601 JAS60168.1 KL367687 KFD60198.1 GECZ01017843 JAS51926.1 GBHO01016409 JAG27195.1 GECZ01000150 JAS69619.1 GDHC01004875 JAQ13754.1 GEZM01099971 JAV53080.1 GDHC01010945 JAQ07684.1 GALX01006157 JAB62309.1 DS567555 QLLG01000626 QKXF01000006 RMX62449.1 RQM18873.1 LBMM01027002 KMQ82250.1 DS567745 DS567497 DS566001 DS566383 GEBQ01029584 JAT10393.1 GEZM01078128 GEZM01078127 JAV63002.1 MJFZ01000618 RAW26780.1 GBHO01041007 JAG02597.1 GDHC01009233 JAQ09396.1 DS567087 LBMM01020254 KMQ83327.1 DS568207 LBMM01012672 KMQ86060.1 DS567139 DS566289 KI913645 ETV63694.1 DS568535 GECL01003258 JAP02866.1 DS568262 NCKW01003798 POM75488.1 DS566231 DS567564 AB378753 BAG68376.1 GEZM01093140 JAV56327.1 DS566082 LBMM01011114 KMQ87021.1 KL367655 KFD60665.1 ABLF02066511 LWTN01000174 ORU94050.1 DS566242 DS566098 BABH01033959 BABH01033960 KQ483738 KYP42039.1 DS567717 GBHO01019280 GBHO01019277 GBHO01019272 GBHO01014987 JAG24324.1 JAG24327.1 JAG24332.1 JAG28617.1 DS568300 KQ971338 KYB28117.1 DS566148 DS566071 NCKW01000038 POM81641.1 DS567770 KN538522 KHJ39825.1 KL367684 KFD60227.1 GEZM01090252 JAV57314.1 KQ483414 KYP53356.1 DS566011 AAZX01006115 AAZX01021021 DS568418 CM003604 KYP72919.1 OIVN01002268 SPD02134.1 KQ483498 KYP48513.1 LYXU01000014 OBS17533.1 CM002870 KFK41513.1 DS567227 ABLF02066512 JXUM01122536 KQ566615 KXJ69965.1 GEZM01048830 JAV76370.1 DS566328 DS568148 NCKW01020291 POM58281.1 OIVN01002376 SPD03107.1 NCKW01004050 POM75106.1 GEZM01002084 JAV97376.1 GBHO01028636 GBHO01028635 JAG14968.1 JAG14969.1 DS566528
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A0A9YBR9
A0A0A9Z091
A0A0A9ZF63
A0A0A9WDS7
A0A0A9W6J9
A0A0J7KC73
+ More
A0A329RKZ4 A0A0A9WDA6 A0A0J7K689 A0A0A9X3Z4 A0A2P4X1D7 A0A1B6GCK5 A0A085MSK2 A0A1B6FNY3 A0A0A9YCF4 A0A1B6H4J4 A0A146M1T4 A0A1Y1JZ65 A0A146LHR9 V5GE66 H3H8R6 A0A3M6V7W3 A0A0J7MNS8 H3GB86 H3H8M0 H3G8C4 H3H5N2 A0A1B6KG29 A0A1Y1KNM9 A0A329RR24 A0A0A9W2F2 A0A146LRM9 H3GB40 A0A0J7JZ17 H3H9X3 A0A0J7N098 H3GB42 H3GAY4 W4F9V3 H3GBA8 A0A0V0G477 H3G6C9 A0A0N5E2R3 A0A2P4YCF8 H3GAW7 H3H8S0 B3Y003 A0A1Y1KC03 A0A1X7U3D2 H3GZJ4 A0A0J7K9V7 A0A085MTW9 A0A1X7STY0 X1WST7 A0A1X1QS81 H3GAW9 H3H1B3 H9JF18 A0A151RHK1 A0A1X7SRH6 H3H922 A0A0A9XUC7 H3HA29 A0A139WJX6 H3GAT9 H3GY81 A0A2P4YV10 H3H949 A0A0B1PJ57 A0A085MSN1 A0A1Y1K7G3 A0A151SEX3 H3GIF1 K7JAV0 H3HAB2 A0A151U0S6 A0A2N9GS71 A0A151S124 A0A1B8AAN0 A0A087HHB1 H3GB53 X1WMK8 A0A182HAB6 A0A1Y1LVN7 H3H5F5 H3H9T4 A0A2P4WYA4 A0A3Q7HLK4 A0A2N9GKY6 A0A2P4YBB4 A0A1Y1NM77 A0A0A9X817 H3H6A9
A0A329RKZ4 A0A0A9WDA6 A0A0J7K689 A0A0A9X3Z4 A0A2P4X1D7 A0A1B6GCK5 A0A085MSK2 A0A1B6FNY3 A0A0A9YCF4 A0A1B6H4J4 A0A146M1T4 A0A1Y1JZ65 A0A146LHR9 V5GE66 H3H8R6 A0A3M6V7W3 A0A0J7MNS8 H3GB86 H3H8M0 H3G8C4 H3H5N2 A0A1B6KG29 A0A1Y1KNM9 A0A329RR24 A0A0A9W2F2 A0A146LRM9 H3GB40 A0A0J7JZ17 H3H9X3 A0A0J7N098 H3GB42 H3GAY4 W4F9V3 H3GBA8 A0A0V0G477 H3G6C9 A0A0N5E2R3 A0A2P4YCF8 H3GAW7 H3H8S0 B3Y003 A0A1Y1KC03 A0A1X7U3D2 H3GZJ4 A0A0J7K9V7 A0A085MTW9 A0A1X7STY0 X1WST7 A0A1X1QS81 H3GAW9 H3H1B3 H9JF18 A0A151RHK1 A0A1X7SRH6 H3H922 A0A0A9XUC7 H3HA29 A0A139WJX6 H3GAT9 H3GY81 A0A2P4YV10 H3H949 A0A0B1PJ57 A0A085MSN1 A0A1Y1K7G3 A0A151SEX3 H3GIF1 K7JAV0 H3HAB2 A0A151U0S6 A0A2N9GS71 A0A151S124 A0A1B8AAN0 A0A087HHB1 H3GB53 X1WMK8 A0A182HAB6 A0A1Y1LVN7 H3H5F5 H3H9T4 A0A2P4WYA4 A0A3Q7HLK4 A0A2N9GKY6 A0A2P4YBB4 A0A1Y1NM77 A0A0A9X817 H3H6A9
Ontologies
PANTHER
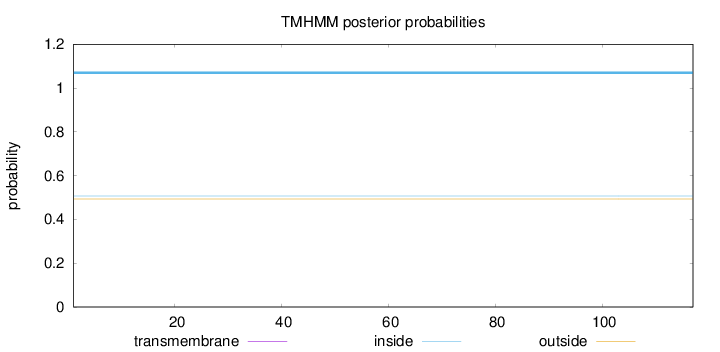
Topology
Length:
117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.50726
inside
1 - 117
Population Genetic Test Statistics
Pi
3.122597
Theta
11.764491
Tajima's D
-2.038829
CLR
0
CSRT
0.0104494775261237
Interpretation
Uncertain
