Pre Gene Modal
BGIBMGA007169
Annotation
ras-like_protein_1_[Bombyx_mori]
Full name
Ras-like protein 1
Alternative Name
Dmras85D
Location in the cell
Cytoplasmic Reliability : 3.417
Sequence
CDS
ATGACCGAGTACAAATTGGTGGTTGTGGGTGCGGGAGGCGTTGGTAAATCCGCATTGACTATACAGCTGATACAGAATCACTTCGTGGACGAATACGATCCCACGATAGAGGATTCGTACCGTAAACAAGTCGTGATCGACGGTGAGACGTGTTTACTCGACATCCTGGACACGGCTGGCCAGGAAGAGTATTCGGCGATGAGAGACCAATACATGCGGACCGGGGAAGGATTCTTGCTGGTATTCGCCGTCAACAGTGCTAAAAGTTTCGAAGACATTGGCTCGTATCGCGAGCAGATCAAGCGAGTGAAGGACGCGGAAGAGGTGCCCATGGTACTTGTGGGCAACAAGTGCGACCTACAGTCGTGGGCCGTAGACATGGCGCGAGCACGAGAGGTGGCGCAAAGCTATAATGTTCCGTTCGTCGAAACATCGGCCAAAACACGCATGGGAGTTGACGACGCTTTCTACACGCTCGTGAGGGAGATACGCAAAGATAAAGTCAGCCGAGACAAGAAATTCAAAGGGAAGAAACCACGTCACGTTCACAAAATAATTAAAGATTGCACCCTTTTTTAA
Protein
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAGQEEYSAMRDQYMRTGEGFLLVFAVNSAKSFEDIGSYREQIKRVKDAEEVPMVLVGNKCDLQSWAVDMARAREVAQSYNVPFVETSAKTRMGVDDAFYTLVREIRKDKVSRDKKFKGKKPRHVHKIIKDCTLF
Summary
Description
Ras proteins bind GDP/GTP and possess intrinsic GTPase activity. Plays a role in eye development by regulating cell growth, survival of postmitotic ommatidial cells and differentiation of photoreceptor cells. During larval development, mediates Ptth/tor signaling leading to the production of ecdysone, a hormone required for the initiation of metamorphosis.
Ras proteins bind GDP/GTP and possess intrinsic GTPase activity (By similarity). Plays a role in eye development by regulating cell growth, survival of postmitotic ommatidial cells and differentiation of photoreceptor cells (PubMed:11290305). During larval development, mediates Ptth/tor signaling leading to the production of ecdysone, a hormone required for the initiation of metamorphosis (PubMed:19965758).
Ras proteins bind GDP/GTP and possess intrinsic GTPase activity (By similarity). Plays a role in eye development by regulating cell growth, survival of postmitotic ommatidial cells and differentiation of photoreceptor cells (PubMed:11290305). During larval development, mediates Ptth/tor signaling leading to the production of ecdysone, a hormone required for the initiation of metamorphosis (PubMed:19965758).
Similarity
Belongs to the small GTPase superfamily. Ras family.
Keywords
Cell membrane
Complete proteome
GTP-binding
Lipoprotein
Membrane
Methylation
Nucleotide-binding
Prenylation
Reference proteome
Feature
chain Ras-like protein 1
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
Q5KT36
A0A212FCP0
A0A0N1I665
E1AXG7
A0A0K8TML9
A0A2W1BNR1
+ More
A0A2A4JWG2 A0A2H1V4W3 A0A437AWZ2 A0A1L8EGY4 T1PFW1 A0A034VYK3 A0A182NMS1 A0A182QRH8 A0A1I8PYA8 A0A1L8DV36 A0A182Y4M9 A0A182LW57 A0A182NZF6 A0A182TCM6 A0A182VM02 A0A182TSC9 A0A182VRX6 A0A084W105 A0A182X368 A0A182IR05 A0A182LQJ2 Q7QK91 A0A182HZJ5 A0A286RSD1 A0A1W4X007 A0A0K8WGR4 W8BVW0 A0A1A9XU59 A0A1B0AVL1 A0A1B0A3K2 D3TQ15 A0A0L0BR78 A0A336LS28 A0A336K696 A0A336LPE2 T1DIT9 Q16N58 A0A023EJ57 A0A1Y1LDW3 U5EY41 A0A336K410 A0A182RH15 A0A0T6AT07 A0A1Q3F0D0 B0WFI4 A0A2A3ER73 A0A088AUS1 K7IUG8 A0A2P8YZW3 A0A0A1XAM4 A0A0C9QIW7 A0A0M8ZRZ6 W5JB40 A0A182FAZ6 A0A0M4N074 A0A2M4AK70 T1DRK1 A0A2R7W4P7 E2A1F7 A0A154PIR6 A0A2M3ZHN1 A0A151IGY0 F4X8P7 A0A195BAI3 A0A195FCW5 A0A158NBH4 E2C6M4 A0A195DRH7 E9JAU0 A0A3L8DTG8 A0A067QHD4 D6W9R1 A0A0L7QL89 A0A1J1IZH1 A0A224XXJ8 A0A0P4VLU4 A0A023F5P8 T1HWY3 A0A069DNZ5 A0A1S3CVU8 A0A291NZI5 A0A291NZD2 A0A291NZB1 A0A291NZ71 A0A291NZJ7 A0A0Q9WNG4 B4LY29 B4PUP5 P83831 B4HKC7 P08646 P83832 B3NZR4 B3M185
A0A2A4JWG2 A0A2H1V4W3 A0A437AWZ2 A0A1L8EGY4 T1PFW1 A0A034VYK3 A0A182NMS1 A0A182QRH8 A0A1I8PYA8 A0A1L8DV36 A0A182Y4M9 A0A182LW57 A0A182NZF6 A0A182TCM6 A0A182VM02 A0A182TSC9 A0A182VRX6 A0A084W105 A0A182X368 A0A182IR05 A0A182LQJ2 Q7QK91 A0A182HZJ5 A0A286RSD1 A0A1W4X007 A0A0K8WGR4 W8BVW0 A0A1A9XU59 A0A1B0AVL1 A0A1B0A3K2 D3TQ15 A0A0L0BR78 A0A336LS28 A0A336K696 A0A336LPE2 T1DIT9 Q16N58 A0A023EJ57 A0A1Y1LDW3 U5EY41 A0A336K410 A0A182RH15 A0A0T6AT07 A0A1Q3F0D0 B0WFI4 A0A2A3ER73 A0A088AUS1 K7IUG8 A0A2P8YZW3 A0A0A1XAM4 A0A0C9QIW7 A0A0M8ZRZ6 W5JB40 A0A182FAZ6 A0A0M4N074 A0A2M4AK70 T1DRK1 A0A2R7W4P7 E2A1F7 A0A154PIR6 A0A2M3ZHN1 A0A151IGY0 F4X8P7 A0A195BAI3 A0A195FCW5 A0A158NBH4 E2C6M4 A0A195DRH7 E9JAU0 A0A3L8DTG8 A0A067QHD4 D6W9R1 A0A0L7QL89 A0A1J1IZH1 A0A224XXJ8 A0A0P4VLU4 A0A023F5P8 T1HWY3 A0A069DNZ5 A0A1S3CVU8 A0A291NZI5 A0A291NZD2 A0A291NZB1 A0A291NZ71 A0A291NZJ7 A0A0Q9WNG4 B4LY29 B4PUP5 P83831 B4HKC7 P08646 P83832 B3NZR4 B3M185
Pubmed
19121390
19946625
22118469
26354079
21167168
26369729
+ More
28756777 25315136 25348373 25244985 24438588 20966253 12364791 14747013 17210077 24495485 20353571 26108605 24330624 17510324 24945155 28004739 20075255 29403074 25830018 20920257 23761445 20798317 21719571 21347285 21282665 30249741 24845553 18362917 19820115 27129103 25474469 26334808 17994087 18057021 10552039 14762063 3110012 6430564 10731132 12537572 12537569 7873789 11290305 18503409 19965758
28756777 25315136 25348373 25244985 24438588 20966253 12364791 14747013 17210077 24495485 20353571 26108605 24330624 17510324 24945155 28004739 20075255 29403074 25830018 20920257 23761445 20798317 21719571 21347285 21282665 30249741 24845553 18362917 19820115 27129103 25474469 26334808 17994087 18057021 10552039 14762063 3110012 6430564 10731132 12537572 12537569 7873789 11290305 18503409 19965758
EMBL
BABH01031555
BABH01031556
BABH01031557
AB176555
BAD83777.1
AGBW02009157
+ More
OWR51502.1 KQ460882 KPJ11484.1 HQ026487 ADM25851.1 GDAI01002185 JAI15418.1 KZ150005 PZC75245.1 NWSH01000508 PCG75974.1 ODYU01000681 SOQ35867.1 RSAL01000317 RVE42590.1 GFDG01000894 JAV17905.1 KA647646 AFP62275.1 GAKP01012335 GAKP01012334 JAC46618.1 AXCN02001208 GFDF01003787 JAV10297.1 AXCM01001303 ATLV01019177 KE525264 KFB43899.1 AXCP01001861 AAAB01008799 EAA03664.4 APCN01001981 KX833970 ASW21098.1 GDHF01008207 GDHF01002042 JAI44107.1 JAI50272.1 GAMC01003188 JAC03368.1 JXJN01004286 CCAG010002237 EZ423517 ADD19793.1 JRES01001582 KNC21734.1 UFQS01000097 UFQT01000097 SSW99391.1 SSX19771.1 SSW99392.1 SSX19772.1 SSW99390.1 SSX19770.1 GALA01000761 JAA94091.1 CH477837 EAT35784.1 GAPW01004744 GEHC01000769 JAC08854.1 JAV46876.1 GEZM01058185 JAV71813.1 GANO01000619 JAB59252.1 SSW99389.1 SSX19769.1 LJIG01022888 KRT78212.1 GFDL01014039 JAV21006.1 DS231918 EDS26252.1 KZ288192 PBC34190.1 AAZX01004436 PYGN01000262 PSN49787.1 GBXI01006564 GBXI01000025 JAD07728.1 JAD14267.1 GBYB01014677 GBYB01014678 GBYB01014679 JAG84444.1 JAG84445.1 JAG84446.1 KQ435883 KOX69747.1 ADMH02002017 GGFL01001270 ETN59979.1 MBW65448.1 KR075835 ALE20554.1 GGFK01007833 MBW41154.1 GAMD01000600 JAB00991.1 KK854327 PTY14693.1 GL435766 EFN72630.1 KQ434926 KZC11693.1 GGFM01007305 MBW28056.1 KQ977661 KYN00688.1 GL888932 EGI57316.1 KQ976540 KYM81242.1 KQ981673 KYN38231.1 ADTU01011108 GL453160 EFN76426.1 KQ980581 KYN15412.1 GL770679 EFZ10058.1 QOIP01000004 RLU23626.1 KK853392 KDR07913.1 KQ971312 EEZ98533.1 KQ414934 KOC59296.1 CVRI01000064 CRL05543.1 GFTR01003164 JAW13262.1 GDKW01000511 JAI56084.1 GBBI01002217 JAC16495.1 ACPB03012299 GBGD01003096 JAC85793.1 KY198393 ATJ26462.1 KY198390 ATJ26459.1 KY198391 ATJ26460.1 KY198389 ATJ26458.1 KY198392 ATJ26461.1 KY198394 KY198395 KY198396 KY198397 KY198398 CH940650 ATJ26463.1 ATJ26464.1 ATJ26465.1 ATJ26466.1 ATJ26467.1 KRF82977.1 EDW66895.1 CM000160 EDW96662.1 AF186649 CM000364 AY459550 AAF15515.1 CH480815 EDW42877.1 M16429 M16123 M16428 K01960 AF186648 AE014297 AY089541 AY094888 X73219 AF186650 AAF15516.1 CH954181 EDV49774.1 CH902617 EDV44355.1
OWR51502.1 KQ460882 KPJ11484.1 HQ026487 ADM25851.1 GDAI01002185 JAI15418.1 KZ150005 PZC75245.1 NWSH01000508 PCG75974.1 ODYU01000681 SOQ35867.1 RSAL01000317 RVE42590.1 GFDG01000894 JAV17905.1 KA647646 AFP62275.1 GAKP01012335 GAKP01012334 JAC46618.1 AXCN02001208 GFDF01003787 JAV10297.1 AXCM01001303 ATLV01019177 KE525264 KFB43899.1 AXCP01001861 AAAB01008799 EAA03664.4 APCN01001981 KX833970 ASW21098.1 GDHF01008207 GDHF01002042 JAI44107.1 JAI50272.1 GAMC01003188 JAC03368.1 JXJN01004286 CCAG010002237 EZ423517 ADD19793.1 JRES01001582 KNC21734.1 UFQS01000097 UFQT01000097 SSW99391.1 SSX19771.1 SSW99392.1 SSX19772.1 SSW99390.1 SSX19770.1 GALA01000761 JAA94091.1 CH477837 EAT35784.1 GAPW01004744 GEHC01000769 JAC08854.1 JAV46876.1 GEZM01058185 JAV71813.1 GANO01000619 JAB59252.1 SSW99389.1 SSX19769.1 LJIG01022888 KRT78212.1 GFDL01014039 JAV21006.1 DS231918 EDS26252.1 KZ288192 PBC34190.1 AAZX01004436 PYGN01000262 PSN49787.1 GBXI01006564 GBXI01000025 JAD07728.1 JAD14267.1 GBYB01014677 GBYB01014678 GBYB01014679 JAG84444.1 JAG84445.1 JAG84446.1 KQ435883 KOX69747.1 ADMH02002017 GGFL01001270 ETN59979.1 MBW65448.1 KR075835 ALE20554.1 GGFK01007833 MBW41154.1 GAMD01000600 JAB00991.1 KK854327 PTY14693.1 GL435766 EFN72630.1 KQ434926 KZC11693.1 GGFM01007305 MBW28056.1 KQ977661 KYN00688.1 GL888932 EGI57316.1 KQ976540 KYM81242.1 KQ981673 KYN38231.1 ADTU01011108 GL453160 EFN76426.1 KQ980581 KYN15412.1 GL770679 EFZ10058.1 QOIP01000004 RLU23626.1 KK853392 KDR07913.1 KQ971312 EEZ98533.1 KQ414934 KOC59296.1 CVRI01000064 CRL05543.1 GFTR01003164 JAW13262.1 GDKW01000511 JAI56084.1 GBBI01002217 JAC16495.1 ACPB03012299 GBGD01003096 JAC85793.1 KY198393 ATJ26462.1 KY198390 ATJ26459.1 KY198391 ATJ26460.1 KY198389 ATJ26458.1 KY198392 ATJ26461.1 KY198394 KY198395 KY198396 KY198397 KY198398 CH940650 ATJ26463.1 ATJ26464.1 ATJ26465.1 ATJ26466.1 ATJ26467.1 KRF82977.1 EDW66895.1 CM000160 EDW96662.1 AF186649 CM000364 AY459550 AAF15515.1 CH480815 EDW42877.1 M16429 M16123 M16428 K01960 AF186648 AE014297 AY089541 AY094888 X73219 AF186650 AAF15516.1 CH954181 EDV49774.1 CH902617 EDV44355.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000218220
UP000283053
UP000095301
+ More
UP000075884 UP000075886 UP000095300 UP000076408 UP000075883 UP000075885 UP000075901 UP000075903 UP000075902 UP000075920 UP000030765 UP000076407 UP000075880 UP000075882 UP000007062 UP000075840 UP000192223 UP000092443 UP000092460 UP000092445 UP000092444 UP000037069 UP000008820 UP000075900 UP000002320 UP000242457 UP000005203 UP000002358 UP000245037 UP000053105 UP000000673 UP000069272 UP000000311 UP000076502 UP000078542 UP000007755 UP000078540 UP000078541 UP000005205 UP000008237 UP000078492 UP000279307 UP000027135 UP000007266 UP000053825 UP000183832 UP000015103 UP000079169 UP000008792 UP000002282 UP000000304 UP000001292 UP000000803 UP000008711 UP000007801
UP000075884 UP000075886 UP000095300 UP000076408 UP000075883 UP000075885 UP000075901 UP000075903 UP000075902 UP000075920 UP000030765 UP000076407 UP000075880 UP000075882 UP000007062 UP000075840 UP000192223 UP000092443 UP000092460 UP000092445 UP000092444 UP000037069 UP000008820 UP000075900 UP000002320 UP000242457 UP000005203 UP000002358 UP000245037 UP000053105 UP000000673 UP000069272 UP000000311 UP000076502 UP000078542 UP000007755 UP000078540 UP000078541 UP000005205 UP000008237 UP000078492 UP000279307 UP000027135 UP000007266 UP000053825 UP000183832 UP000015103 UP000079169 UP000008792 UP000002282 UP000000304 UP000001292 UP000000803 UP000008711 UP000007801
Interpro
ProteinModelPortal
Q5KT36
A0A212FCP0
A0A0N1I665
E1AXG7
A0A0K8TML9
A0A2W1BNR1
+ More
A0A2A4JWG2 A0A2H1V4W3 A0A437AWZ2 A0A1L8EGY4 T1PFW1 A0A034VYK3 A0A182NMS1 A0A182QRH8 A0A1I8PYA8 A0A1L8DV36 A0A182Y4M9 A0A182LW57 A0A182NZF6 A0A182TCM6 A0A182VM02 A0A182TSC9 A0A182VRX6 A0A084W105 A0A182X368 A0A182IR05 A0A182LQJ2 Q7QK91 A0A182HZJ5 A0A286RSD1 A0A1W4X007 A0A0K8WGR4 W8BVW0 A0A1A9XU59 A0A1B0AVL1 A0A1B0A3K2 D3TQ15 A0A0L0BR78 A0A336LS28 A0A336K696 A0A336LPE2 T1DIT9 Q16N58 A0A023EJ57 A0A1Y1LDW3 U5EY41 A0A336K410 A0A182RH15 A0A0T6AT07 A0A1Q3F0D0 B0WFI4 A0A2A3ER73 A0A088AUS1 K7IUG8 A0A2P8YZW3 A0A0A1XAM4 A0A0C9QIW7 A0A0M8ZRZ6 W5JB40 A0A182FAZ6 A0A0M4N074 A0A2M4AK70 T1DRK1 A0A2R7W4P7 E2A1F7 A0A154PIR6 A0A2M3ZHN1 A0A151IGY0 F4X8P7 A0A195BAI3 A0A195FCW5 A0A158NBH4 E2C6M4 A0A195DRH7 E9JAU0 A0A3L8DTG8 A0A067QHD4 D6W9R1 A0A0L7QL89 A0A1J1IZH1 A0A224XXJ8 A0A0P4VLU4 A0A023F5P8 T1HWY3 A0A069DNZ5 A0A1S3CVU8 A0A291NZI5 A0A291NZD2 A0A291NZB1 A0A291NZ71 A0A291NZJ7 A0A0Q9WNG4 B4LY29 B4PUP5 P83831 B4HKC7 P08646 P83832 B3NZR4 B3M185
A0A2A4JWG2 A0A2H1V4W3 A0A437AWZ2 A0A1L8EGY4 T1PFW1 A0A034VYK3 A0A182NMS1 A0A182QRH8 A0A1I8PYA8 A0A1L8DV36 A0A182Y4M9 A0A182LW57 A0A182NZF6 A0A182TCM6 A0A182VM02 A0A182TSC9 A0A182VRX6 A0A084W105 A0A182X368 A0A182IR05 A0A182LQJ2 Q7QK91 A0A182HZJ5 A0A286RSD1 A0A1W4X007 A0A0K8WGR4 W8BVW0 A0A1A9XU59 A0A1B0AVL1 A0A1B0A3K2 D3TQ15 A0A0L0BR78 A0A336LS28 A0A336K696 A0A336LPE2 T1DIT9 Q16N58 A0A023EJ57 A0A1Y1LDW3 U5EY41 A0A336K410 A0A182RH15 A0A0T6AT07 A0A1Q3F0D0 B0WFI4 A0A2A3ER73 A0A088AUS1 K7IUG8 A0A2P8YZW3 A0A0A1XAM4 A0A0C9QIW7 A0A0M8ZRZ6 W5JB40 A0A182FAZ6 A0A0M4N074 A0A2M4AK70 T1DRK1 A0A2R7W4P7 E2A1F7 A0A154PIR6 A0A2M3ZHN1 A0A151IGY0 F4X8P7 A0A195BAI3 A0A195FCW5 A0A158NBH4 E2C6M4 A0A195DRH7 E9JAU0 A0A3L8DTG8 A0A067QHD4 D6W9R1 A0A0L7QL89 A0A1J1IZH1 A0A224XXJ8 A0A0P4VLU4 A0A023F5P8 T1HWY3 A0A069DNZ5 A0A1S3CVU8 A0A291NZI5 A0A291NZD2 A0A291NZB1 A0A291NZ71 A0A291NZJ7 A0A0Q9WNG4 B4LY29 B4PUP5 P83831 B4HKC7 P08646 P83832 B3NZR4 B3M185
PDB
5USJ
E-value=5.77696e-71,
Score=676
Ontologies
PATHWAY
04013
MAPK signaling pathway - fly - Bombyx mori (domestic silkworm)
04068 FoxO signaling pathway - Bombyx mori (domestic silkworm)
04137 Mitophagy - animal - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04150 mTOR signaling pathway - Bombyx mori (domestic silkworm)
04213 Longevity regulating pathway - multiple species - Bombyx mori (domestic silkworm)
04214 Apoptosis - fly - Bombyx mori (domestic silkworm)
04320 Dorso-ventral axis formation - Bombyx mori (domestic silkworm)
04933 AGE-RAGE signaling pathway in diabetic complications - Bombyx mori (domestic silkworm)
04068 FoxO signaling pathway - Bombyx mori (domestic silkworm)
04137 Mitophagy - animal - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04150 mTOR signaling pathway - Bombyx mori (domestic silkworm)
04213 Longevity regulating pathway - multiple species - Bombyx mori (domestic silkworm)
04214 Apoptosis - fly - Bombyx mori (domestic silkworm)
04320 Dorso-ventral axis formation - Bombyx mori (domestic silkworm)
04933 AGE-RAGE signaling pathway in diabetic complications - Bombyx mori (domestic silkworm)
GO
GO:0005525
GO:0016020
GO:0007165
GO:0003924
GO:0007265
GO:0019003
GO:0005886
GO:0007173
GO:0003677
GO:0006260
GO:0005524
GO:0036335
GO:0008340
GO:0046534
GO:0009267
GO:0002168
GO:0008293
GO:0048865
GO:0007455
GO:0010629
GO:0072089
GO:0007479
GO:0043703
GO:0072002
GO:0007430
GO:0008594
GO:0040014
GO:0048010
GO:0007395
GO:0035088
GO:0060438
GO:0051607
GO:0007427
GO:0007474
GO:0045500
GO:0030381
GO:0035208
GO:2001234
GO:0070374
GO:0007362
GO:0007422
GO:0016242
GO:0035169
GO:0008586
GO:0016318
GO:0007298
GO:0000165
GO:0008543
GO:0007309
GO:0001710
GO:0046673
GO:0007426
GO:0045793
GO:0048626
GO:1904263
GO:0035099
GO:0035170
GO:0001709
GO:0008361
GO:0040008
GO:0007465
GO:0007369
GO:0007391
GO:0008283
GO:0046530
GO:0007552
GO:0042981
GO:0007472
GO:0007476
GO:0007623
GO:0043066
GO:0008284
GO:0048749
GO:0048863
GO:0010507
GO:0042461
GO:0007264
GO:0005515
GO:0004003
GO:0005634
GO:0006289
GO:0016286
PANTHER
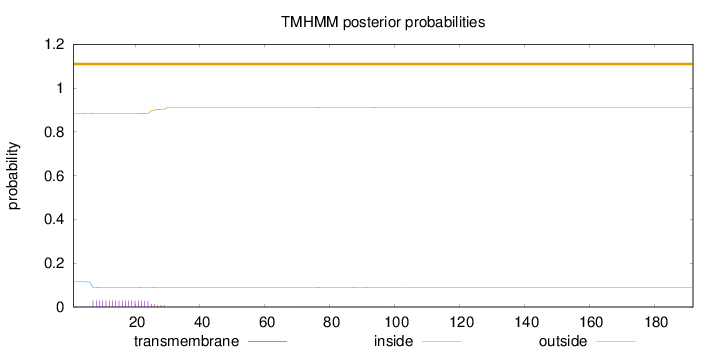
Topology
Subcellular location
Cell membrane
Length:
192
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.59425
Exp number, first 60 AAs:
0.57744
Total prob of N-in:
0.11669
outside
1 - 192
Population Genetic Test Statistics
Pi
3.858139
Theta
15.655732
Tajima's D
-0.990226
CLR
0.509741
CSRT
0.138843057847108
Interpretation
Uncertain
