Pre Gene Modal
BGIBMGA007194
Annotation
PREDICTED:_lon_protease_homolog?_mitochondrial_isoform_X2_[Bombyx_mori]
Full name
Lon protease homolog, mitochondrial
+ More
Lon protease homolog
Lon protease homolog
Location in the cell
Mitochondrial Reliability : 2.069
Sequence
CDS
ATGTCGGGGTACATCGCGGAGGAGAAACTGGCCATAGCGCAGCAGTACCTGGTGCCGACGGCACGGCAGAACTGTGGGCTCTCCCAGCAGCAGCTGGCGGTGTCGGAGCCGGCGCTGCTGGCGCTCGTGCGCTCCTACTGCCGCGAGAGCGGAGTCCGAAACCTGCAGAAGCACATTGAAAAGATAGCCCGCAAGGTGGCTTATAAAATAGTGAAGAAGGAAACGGAAGAGGTCAACGTCACGGAAGAGAACCTGTCGGATTTGGTCGGCAAGCCTAGCTTCAAGCACGACCGCATGTATGACGTCACCCCGCCCGGCGTCATCATGGGACTAGCCTGGACCGCTATGGGGGGCAGCACACTGTACATAGAGACTGCACTGAGGAATAGCACACAGGACGAGAAGTCGCCCTTCGGCTCGCTGGAGCTGACCGGCCACCTCGGGGACGTCATGAAGGAGTCCGCTCGCATCGCGCTCACCGTCGCTAGGAATTACCTCGCCACCCACCACCCTGACAATAAGTTCTTGAACACGAGCCACCTCCACCTGCACGTGCCGGAGGGCGCCACCCCGAAGGACGGGCCGTCGGCCGGGGTCACCATCACGTCGGCGCTGCTGTCGCTGGCTCTGGGCAGGGCTCCGCGGGCCGACCTCGCCATGACGGGGGAGGTGTCCCTCACCGGTCGGGTGCTGCCCGTCGGCGGCATCAAGGAGAAGATTATAGCCGCGAAACGCGTGGGCGTGAAGTGCGTGATCCTTCCGGAGGAGAACAGGCGGGACTACGAGGACCTGCAGTCCTTCATCAAGCAAGACATAGACGTGCACTTCGTCAGCACGTACGACGACGTGTTCAGGGTCGCCTTTGGAGAGCAGCAGATATTCGAGGCCGACCACTCATCCACATCGGAGCTTTAA
Protein
MSGYIAEEKLAIAQQYLVPTARQNCGLSQQQLAVSEPALLALVRSYCRESGVRNLQKHIEKIARKVAYKIVKKETEEVNVTEENLSDLVGKPSFKHDRMYDVTPPGVIMGLAWTAMGGSTLYIETALRNSTQDEKSPFGSLELTGHLGDVMKESARIALTVARNYLATHHPDNKFLNTSHLHLHVPEGATPKDGPSAGVTITSALLSLALGRAPRADLAMTGEVSLTGRVLPVGGIKEKIIAAKRVGVKCVILPEENRRDYEDLQSFIKQDIDVHFVSTYDDVFRVAFGEQQIFEADHSSTSEL
Summary
Description
ATP-dependent serine protease that mediates the selective degradation of misfolded, unassembled or oxidatively damaged polypeptides as well as certain short-lived regulatory proteins in the mitochondrial matrix. May also have a chaperone function in the assembly of inner membrane protein complexes. Participates in the regulation of mitochondrial gene expression and in the maintenance of the integrity of the mitochondrial genome. Binds to mitochondrial DNA in a site-specific manner.
Catalytic Activity
Hydrolysis of proteins in presence of ATP.
Subunit
Homohexamer or homoheptamer. Organized in a ring with a central cavity.
Similarity
Belongs to the peptidase S16 family.
Uniprot
H9JCE9
A0A2H1V4Z6
A0A194QM00
A0A2A4JX53
A0A3S2LR09
A0A212FCQ2
+ More
A0A2J7RJS7 A0A2J7RJT0 A0A0N0PBV6 N6T429 A0A158NUV2 E0VJG4 A0A195B8I8 A0A1B6MLC6 A0A1B6MLV5 A0A1L8DTJ0 A0A1B6HV54 U5EI28 A0A151IHZ3 A0A067QTG5 E2AFM0 K7IWC8 A0A182U9Z5 A0A1Y1LEI3 B0WP21 Q176B8 A0A1B0GKA2 A0A2F0BEE6 A0A2M4BEK3 A0A232F8W0 A0A182G8C0 A0A182JXX0 A0A182PSC1 E2BX97 A0A0N1ITX4 A0A182NDM9 A0A195FVU6 A0A1L8DTV2 A0A182UWV5 A0A0J7NG97 A0A151WNM8 A0A182Q276 A0A1Q3FX66 A0A182T7M0 A0A1Q3FWQ0 A0A2M4BCF8 A0A1B6IB05 A0A1B6HJ87 A0A2M4BCG1 D6W9J3 F4WNM2 A0A1Y1L8P7 U4UB30 A0A0A9X8U9 A0A2M4BCC6 A0A0P4VU21 A0A1Y1LG27 A0A182LNA9 A0A0K8S447 T1I5V3 A0A146MED5 A0A2P4SSW0 A0A0V0G6L8 A0A182HXB1 A0A182JL79 A0A182FM56 A0A1S4H431 A0A069DWP6 A0A182X851 A0A2M4A9T4 A0A224XBA6 A0A2M4A9L1 A0A1B6BZM9 Q7QDW1 A0A182RJQ2 A0A026WSQ1 A0A3L8DH63 T1PPM5 A0A182M6P9 Q56A16 W5JW89 A0A182YIQ7 A0A1B6H1P4 A0A1B6G104 A0A310SD14 A0A0L7R5Z5 A0A084VDC5 A0A2A3EKF6 A0A183U0V4 A0A3P7EVZ7 A0A088A462 A0A336LJ53 A0A023F4V0 A0A1I8M1H4 A0A1I8M1H7 F1L992 A0A336LMV6 A0A2J8JRE1 A0A1U7SFG0 A0A1I8PJS4
A0A2J7RJS7 A0A2J7RJT0 A0A0N0PBV6 N6T429 A0A158NUV2 E0VJG4 A0A195B8I8 A0A1B6MLC6 A0A1B6MLV5 A0A1L8DTJ0 A0A1B6HV54 U5EI28 A0A151IHZ3 A0A067QTG5 E2AFM0 K7IWC8 A0A182U9Z5 A0A1Y1LEI3 B0WP21 Q176B8 A0A1B0GKA2 A0A2F0BEE6 A0A2M4BEK3 A0A232F8W0 A0A182G8C0 A0A182JXX0 A0A182PSC1 E2BX97 A0A0N1ITX4 A0A182NDM9 A0A195FVU6 A0A1L8DTV2 A0A182UWV5 A0A0J7NG97 A0A151WNM8 A0A182Q276 A0A1Q3FX66 A0A182T7M0 A0A1Q3FWQ0 A0A2M4BCF8 A0A1B6IB05 A0A1B6HJ87 A0A2M4BCG1 D6W9J3 F4WNM2 A0A1Y1L8P7 U4UB30 A0A0A9X8U9 A0A2M4BCC6 A0A0P4VU21 A0A1Y1LG27 A0A182LNA9 A0A0K8S447 T1I5V3 A0A146MED5 A0A2P4SSW0 A0A0V0G6L8 A0A182HXB1 A0A182JL79 A0A182FM56 A0A1S4H431 A0A069DWP6 A0A182X851 A0A2M4A9T4 A0A224XBA6 A0A2M4A9L1 A0A1B6BZM9 Q7QDW1 A0A182RJQ2 A0A026WSQ1 A0A3L8DH63 T1PPM5 A0A182M6P9 Q56A16 W5JW89 A0A182YIQ7 A0A1B6H1P4 A0A1B6G104 A0A310SD14 A0A0L7R5Z5 A0A084VDC5 A0A2A3EKF6 A0A183U0V4 A0A3P7EVZ7 A0A088A462 A0A336LJ53 A0A023F4V0 A0A1I8M1H4 A0A1I8M1H7 F1L992 A0A336LMV6 A0A2J8JRE1 A0A1U7SFG0 A0A1I8PJS4
EC Number
3.4.21.53
3.4.21.-
3.4.21.-
Pubmed
EMBL
BABH01031552
ODYU01000681
SOQ35869.1
KQ458981
KPJ04481.1
NWSH01000508
+ More
PCG75972.1 RSAL01000317 RVE42591.1 AGBW02009157 OWR51500.1 NEVH01002988 PNF41086.1 PNF41087.1 KQ460882 KPJ11486.1 APGK01044427 KB741026 ENN74914.1 ADTU01002868 ADTU01002869 DS235222 EEB13520.1 KQ976565 KYM80509.1 GEBQ01003263 JAT36714.1 GEBQ01003047 JAT36930.1 GFDF01004312 JAV09772.1 GECU01029180 JAS78526.1 GANO01002794 JAB57077.1 KQ977545 KYN02031.1 KK853510 KDR07078.1 GL439118 EFN67771.1 GEZM01062186 JAV69966.1 DS232018 EDS32048.1 CH477391 EAT41940.1 AJWK01028599 NTJE010156281 MBW01356.1 GGFJ01002334 MBW51475.1 NNAY01000634 OXU27284.1 JXUM01047603 JXUM01047604 JXUM01047605 JXUM01047606 JXUM01047607 JXUM01047608 JXUM01047609 JXUM01047610 KQ561527 KXJ78274.1 GL451230 EFN79680.1 KQ435728 KOX77752.1 KQ981215 KYN44546.1 GFDF01004359 JAV09725.1 LBMM01005390 KMQ91560.1 KQ982918 KYQ49295.1 AXCN02001747 GFDL01003032 JAV32013.1 GFDL01003036 JAV32009.1 GGFJ01001562 MBW50703.1 GECU01023582 JAS84124.1 GECU01032947 JAS74759.1 GGFJ01001561 MBW50702.1 KQ971312 EEZ98145.1 GL888237 EGI64215.1 GEZM01062187 JAV69964.1 KB631924 ERL87165.1 GBHO01026487 JAG17117.1 GGFJ01001563 MBW50704.1 GDKW01002263 JAI54332.1 GEZM01062185 JAV69967.1 GBRD01017766 JAG48061.1 ACPB03008304 GDHC01006234 GDHC01000471 JAQ12395.1 JAQ18158.1 PPHD01024931 POI27196.1 GECL01002438 JAP03686.1 APCN01005172 AAAB01008849 GBGD01000421 JAC88468.1 GGFK01004243 MBW37564.1 GFTR01008212 JAW08214.1 GGFK01004175 MBW37496.1 GEDC01030575 JAS06723.1 EAA07151.4 KK107109 EZA59062.1 QOIP01000008 RLU19522.1 KA650140 AFP64769.1 AXCM01000213 BC092212 AAH92212.1 ADMH02000098 ETN67828.1 GECZ01001202 JAS68567.1 GECZ01013663 JAS56106.1 KQ762962 OAD55184.1 KQ414648 KOC66312.1 ATLV01011253 KE524658 KFB35969.1 KZ288219 PBC32293.1 UYWY01001958 VDM27510.1 UFQT01000027 SSX18112.1 GBBI01002669 JAC16043.1 JI174384 ADY46696.1 SSX18111.1 NBAG03000432 PNI25343.1
PCG75972.1 RSAL01000317 RVE42591.1 AGBW02009157 OWR51500.1 NEVH01002988 PNF41086.1 PNF41087.1 KQ460882 KPJ11486.1 APGK01044427 KB741026 ENN74914.1 ADTU01002868 ADTU01002869 DS235222 EEB13520.1 KQ976565 KYM80509.1 GEBQ01003263 JAT36714.1 GEBQ01003047 JAT36930.1 GFDF01004312 JAV09772.1 GECU01029180 JAS78526.1 GANO01002794 JAB57077.1 KQ977545 KYN02031.1 KK853510 KDR07078.1 GL439118 EFN67771.1 GEZM01062186 JAV69966.1 DS232018 EDS32048.1 CH477391 EAT41940.1 AJWK01028599 NTJE010156281 MBW01356.1 GGFJ01002334 MBW51475.1 NNAY01000634 OXU27284.1 JXUM01047603 JXUM01047604 JXUM01047605 JXUM01047606 JXUM01047607 JXUM01047608 JXUM01047609 JXUM01047610 KQ561527 KXJ78274.1 GL451230 EFN79680.1 KQ435728 KOX77752.1 KQ981215 KYN44546.1 GFDF01004359 JAV09725.1 LBMM01005390 KMQ91560.1 KQ982918 KYQ49295.1 AXCN02001747 GFDL01003032 JAV32013.1 GFDL01003036 JAV32009.1 GGFJ01001562 MBW50703.1 GECU01023582 JAS84124.1 GECU01032947 JAS74759.1 GGFJ01001561 MBW50702.1 KQ971312 EEZ98145.1 GL888237 EGI64215.1 GEZM01062187 JAV69964.1 KB631924 ERL87165.1 GBHO01026487 JAG17117.1 GGFJ01001563 MBW50704.1 GDKW01002263 JAI54332.1 GEZM01062185 JAV69967.1 GBRD01017766 JAG48061.1 ACPB03008304 GDHC01006234 GDHC01000471 JAQ12395.1 JAQ18158.1 PPHD01024931 POI27196.1 GECL01002438 JAP03686.1 APCN01005172 AAAB01008849 GBGD01000421 JAC88468.1 GGFK01004243 MBW37564.1 GFTR01008212 JAW08214.1 GGFK01004175 MBW37496.1 GEDC01030575 JAS06723.1 EAA07151.4 KK107109 EZA59062.1 QOIP01000008 RLU19522.1 KA650140 AFP64769.1 AXCM01000213 BC092212 AAH92212.1 ADMH02000098 ETN67828.1 GECZ01001202 JAS68567.1 GECZ01013663 JAS56106.1 KQ762962 OAD55184.1 KQ414648 KOC66312.1 ATLV01011253 KE524658 KFB35969.1 KZ288219 PBC32293.1 UYWY01001958 VDM27510.1 UFQT01000027 SSX18112.1 GBBI01002669 JAC16043.1 JI174384 ADY46696.1 SSX18111.1 NBAG03000432 PNI25343.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000283053
UP000007151
UP000235965
+ More
UP000053240 UP000019118 UP000005205 UP000009046 UP000078540 UP000078542 UP000027135 UP000000311 UP000002358 UP000075902 UP000002320 UP000008820 UP000092461 UP000215335 UP000069940 UP000249989 UP000075881 UP000075885 UP000008237 UP000053105 UP000075884 UP000078541 UP000075903 UP000036403 UP000075809 UP000075886 UP000075901 UP000007266 UP000007755 UP000030742 UP000075882 UP000015103 UP000075840 UP000075880 UP000069272 UP000076407 UP000007062 UP000075900 UP000053097 UP000279307 UP000075883 UP000000673 UP000076408 UP000053825 UP000030765 UP000242457 UP000050794 UP000267007 UP000005203 UP000095301 UP000189705 UP000095300
UP000053240 UP000019118 UP000005205 UP000009046 UP000078540 UP000078542 UP000027135 UP000000311 UP000002358 UP000075902 UP000002320 UP000008820 UP000092461 UP000215335 UP000069940 UP000249989 UP000075881 UP000075885 UP000008237 UP000053105 UP000075884 UP000078541 UP000075903 UP000036403 UP000075809 UP000075886 UP000075901 UP000007266 UP000007755 UP000030742 UP000075882 UP000015103 UP000075840 UP000075880 UP000069272 UP000076407 UP000007062 UP000075900 UP000053097 UP000279307 UP000075883 UP000000673 UP000076408 UP000053825 UP000030765 UP000242457 UP000050794 UP000267007 UP000005203 UP000095301 UP000189705 UP000095300
PRIDE
Interpro
IPR008269
Lon_proteolytic
+ More
IPR008268 Peptidase_S16_AS
IPR027065 Lon_Prtase
IPR003593 AAA+_ATPase
IPR015947 PUA-like_sf
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR004815 Lon_bac/euk-typ
IPR027503 Lonm_euk
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR020568 Ribosomal_S5_D2-typ_fold
IPR003111 Lon_substr-bd
IPR004572 Protoporphyrinogen_oxidase
IPR036188 FAD/NAD-bd_sf
IPR002937 Amino_oxidase
IPR008268 Peptidase_S16_AS
IPR027065 Lon_Prtase
IPR003593 AAA+_ATPase
IPR015947 PUA-like_sf
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR004815 Lon_bac/euk-typ
IPR027503 Lonm_euk
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR020568 Ribosomal_S5_D2-typ_fold
IPR003111 Lon_substr-bd
IPR004572 Protoporphyrinogen_oxidase
IPR036188 FAD/NAD-bd_sf
IPR002937 Amino_oxidase
Gene 3D
ProteinModelPortal
H9JCE9
A0A2H1V4Z6
A0A194QM00
A0A2A4JX53
A0A3S2LR09
A0A212FCQ2
+ More
A0A2J7RJS7 A0A2J7RJT0 A0A0N0PBV6 N6T429 A0A158NUV2 E0VJG4 A0A195B8I8 A0A1B6MLC6 A0A1B6MLV5 A0A1L8DTJ0 A0A1B6HV54 U5EI28 A0A151IHZ3 A0A067QTG5 E2AFM0 K7IWC8 A0A182U9Z5 A0A1Y1LEI3 B0WP21 Q176B8 A0A1B0GKA2 A0A2F0BEE6 A0A2M4BEK3 A0A232F8W0 A0A182G8C0 A0A182JXX0 A0A182PSC1 E2BX97 A0A0N1ITX4 A0A182NDM9 A0A195FVU6 A0A1L8DTV2 A0A182UWV5 A0A0J7NG97 A0A151WNM8 A0A182Q276 A0A1Q3FX66 A0A182T7M0 A0A1Q3FWQ0 A0A2M4BCF8 A0A1B6IB05 A0A1B6HJ87 A0A2M4BCG1 D6W9J3 F4WNM2 A0A1Y1L8P7 U4UB30 A0A0A9X8U9 A0A2M4BCC6 A0A0P4VU21 A0A1Y1LG27 A0A182LNA9 A0A0K8S447 T1I5V3 A0A146MED5 A0A2P4SSW0 A0A0V0G6L8 A0A182HXB1 A0A182JL79 A0A182FM56 A0A1S4H431 A0A069DWP6 A0A182X851 A0A2M4A9T4 A0A224XBA6 A0A2M4A9L1 A0A1B6BZM9 Q7QDW1 A0A182RJQ2 A0A026WSQ1 A0A3L8DH63 T1PPM5 A0A182M6P9 Q56A16 W5JW89 A0A182YIQ7 A0A1B6H1P4 A0A1B6G104 A0A310SD14 A0A0L7R5Z5 A0A084VDC5 A0A2A3EKF6 A0A183U0V4 A0A3P7EVZ7 A0A088A462 A0A336LJ53 A0A023F4V0 A0A1I8M1H4 A0A1I8M1H7 F1L992 A0A336LMV6 A0A2J8JRE1 A0A1U7SFG0 A0A1I8PJS4
A0A2J7RJS7 A0A2J7RJT0 A0A0N0PBV6 N6T429 A0A158NUV2 E0VJG4 A0A195B8I8 A0A1B6MLC6 A0A1B6MLV5 A0A1L8DTJ0 A0A1B6HV54 U5EI28 A0A151IHZ3 A0A067QTG5 E2AFM0 K7IWC8 A0A182U9Z5 A0A1Y1LEI3 B0WP21 Q176B8 A0A1B0GKA2 A0A2F0BEE6 A0A2M4BEK3 A0A232F8W0 A0A182G8C0 A0A182JXX0 A0A182PSC1 E2BX97 A0A0N1ITX4 A0A182NDM9 A0A195FVU6 A0A1L8DTV2 A0A182UWV5 A0A0J7NG97 A0A151WNM8 A0A182Q276 A0A1Q3FX66 A0A182T7M0 A0A1Q3FWQ0 A0A2M4BCF8 A0A1B6IB05 A0A1B6HJ87 A0A2M4BCG1 D6W9J3 F4WNM2 A0A1Y1L8P7 U4UB30 A0A0A9X8U9 A0A2M4BCC6 A0A0P4VU21 A0A1Y1LG27 A0A182LNA9 A0A0K8S447 T1I5V3 A0A146MED5 A0A2P4SSW0 A0A0V0G6L8 A0A182HXB1 A0A182JL79 A0A182FM56 A0A1S4H431 A0A069DWP6 A0A182X851 A0A2M4A9T4 A0A224XBA6 A0A2M4A9L1 A0A1B6BZM9 Q7QDW1 A0A182RJQ2 A0A026WSQ1 A0A3L8DH63 T1PPM5 A0A182M6P9 Q56A16 W5JW89 A0A182YIQ7 A0A1B6H1P4 A0A1B6G104 A0A310SD14 A0A0L7R5Z5 A0A084VDC5 A0A2A3EKF6 A0A183U0V4 A0A3P7EVZ7 A0A088A462 A0A336LJ53 A0A023F4V0 A0A1I8M1H4 A0A1I8M1H7 F1L992 A0A336LMV6 A0A2J8JRE1 A0A1U7SFG0 A0A1I8PJS4
PDB
2X36
E-value=4.62426e-72,
Score=688
Ontologies
GO
PANTHER
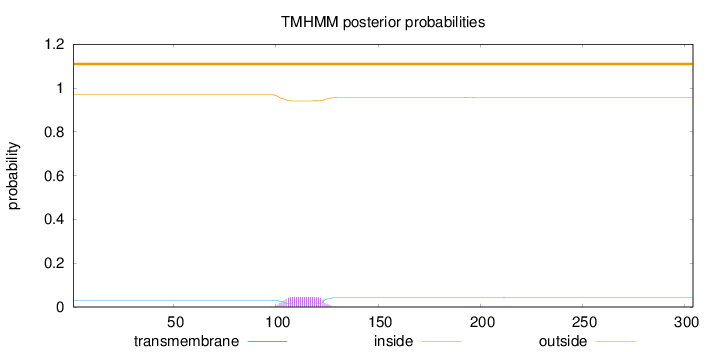
Topology
Subcellular location
Mitochondrion matrix
Length:
304
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.98456
Exp number, first 60 AAs:
0.00033
Total prob of N-in:
0.03063
outside
1 - 304
Population Genetic Test Statistics
Pi
18.464002
Theta
17.73133
Tajima's D
-0.808283
CLR
1.30122
CSRT
0.176041197940103
Interpretation
Uncertain
