Pre Gene Modal
BGIBMGA007171
Annotation
PREDICTED:_probable_tRNA_(guanine(26)-N(2))-dimethyltransferase_isoform_X1_[Bombyx_mori]
Full name
tRNA (guanine(26)-N(2))-dimethyltransferase
+ More
Probable tRNA (guanine(26)-N(2))-dimethyltransferase
Probable tRNA (guanine(26)-N(2))-dimethyltransferase
Alternative Name
tRNA 2,2-dimethylguanosine-26 methyltransferase
tRNA(guanine-26,N(2)-N(2)) methyltransferase
tRNA(m(2,2)G26)dimethyltransferase
tRNA(guanine-26,N(2)-N(2)) methyltransferase
tRNA(m(2,2)G26)dimethyltransferase
Location in the cell
Nuclear Reliability : 3.221
Sequence
CDS
ATGAGAATGGAAACTATATCAAACTTAAAAACTATAAAAGAAGGCCAAGCAGAGATATGTTTAACTACAGAGAAGGTTTTTTACAATCCTGTTCAAGAATTCAACCGTGATCTTAGTATAGCTGTGCTTACGCTGTTTATTGAAGATTATAAAGCGGAAAAGTTGGCACGTTTCGAAAAAAAACAAAAAAAACTGGAAACCGTGCAAGATGAAGAAAGCGGAGGTAATCCAGAGCCAAAAATAACAATCTTAGAAGCACTATCAGCAACAGGCCTCAGAAGCATCCGTTATGCTAAAGAAATACCATATGCTACCAATATCATTGCAAATGATCTCTCAGAACAAGCTGTTGAGACAATTAAGCATAATATTGAACATAACCAGGTGTCACGCATCATAGAAACTAGTCATGATGATGCTTGTATGTTAATGTACAAACATAAACACCCTTCAAAGCGTTTTGCGGCTATTGATCTGGACCCTTATGGATGCCCTTCAATATTCCTCGACTCTGCAGTACAGAGCATTCAAGATGGTGGTTTGCTACTCGTTACAGCAACTGATATGGCAGTTTTAGCCGGCAACTCACCAGAGACATGTTATTGTAAATATGGTGCTGTTAGCTTAAAGACCAAATGCTGTCATGAGATGGCCCTCAGGATAATGCTACAATGTATAGAACAGCATGCAAATCGTTACAGCAGATACATTGTTCCAATTCTTAGTATATCAGCCGATTTCTACATACGTGTTTTTGTCAAAATATACTCTGGTGCCGTGCACTGCAAGAAGACAACAAGCAAGTTGTCAATGGTTTACCACTGCACAGGCTGCGATAACTTGACCCTGCAACCTCTGGGTGGCTTAAAACCGAACCCGTCTGAGAAAAACCCAAACCAAATGAAGGCTTATCTTCCTGCAGCACCACCTATAGGCGAATACTGCACCAACTGCAACCAGAGGCATCATCTAGGCGGTCCAATATGGTCGGCTCCGATACACGACGAATCCTTCGTGTCACGCGTCCTAACGTACGTAGAAGAGAATTCTCAACTCTTCGGAACGGCGAAACGCATTGAAGGCGTCTTGTCTATGGTCCGAGAGGAGTTGAACGACGTCCCTCTGTACTACACCATAGACAAGTTATTCGGTAGAGTGCACTTGCAGACAATGCCGATGTTGATAATGAGATCAGCAATTATTAACGCTGGCTATAAAGTATCTTATTCGCACGCATCGAAAATGTCAGTGAAGACGAACGCTCCACCACAATTGATGTGGGACATCATCAGGACGTGGGAGAAAACACACCCAATCAAGCAGAGCAAACTGGTTTCAGATCCTGTAACGAAGCACATACTATCTCAGGATATAAAGAATAATGTCGATTTGTCGGTGAATGAGCTAGCCAATCCGCTGAGCCGCCGTGACGGCAGGCTTCGCTTCCAGATCAATCCTGCACCGTTCTGGGGACCTGGGTCCAGAGCCGCAGTCAATATCGGCGACGACAAAATGTCGAAGTCTGTTAAAAATCAGAACAAGAACAGGAAGAAAGAAAACAATGTTAAAAGAAAACATTCGGATACAAGTGACGATGATTCACGGAAAAAAGTTGATATGGAATTAAAACATAATGAATAA
Protein
MRMETISNLKTIKEGQAEICLTTEKVFYNPVQEFNRDLSIAVLTLFIEDYKAEKLARFEKKQKKLETVQDEESGGNPEPKITILEALSATGLRSIRYAKEIPYATNIIANDLSEQAVETIKHNIEHNQVSRIIETSHDDACMLMYKHKHPSKRFAAIDLDPYGCPSIFLDSAVQSIQDGGLLLVTATDMAVLAGNSPETCYCKYGAVSLKTKCCHEMALRIMLQCIEQHANRYSRYIVPILSISADFYIRVFVKIYSGAVHCKKTTSKLSMVYHCTGCDNLTLQPLGGLKPNPSEKNPNQMKAYLPAAPPIGEYCTNCNQRHHLGGPIWSAPIHDESFVSRVLTYVEENSQLFGTAKRIEGVLSMVREELNDVPLYYTIDKLFGRVHLQTMPMLIMRSAIINAGYKVSYSHASKMSVKTNAPPQLMWDIIRTWEKTHPIKQSKLVSDPVTKHILSQDIKNNVDLSVNELANPLSRRDGRLRFQINPAPFWGPGSRAAVNIGDDKMSKSVKNQNKNRKKENNVKRKHSDTSDDDSRKKVDMELKHNE
Summary
Description
Dimethylates a single guanine residue at position 26 of most tRNAs using S-adenosyl-L-methionine as donor of the methyl groups.
Catalytic Activity
guanosine(26) in tRNA + 2 S-adenosyl-L-methionine = 2 H(+) + N(2)-dimethylguanosine(26) in tRNA + 2 S-adenosyl-L-homocysteine
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. Trm1 family.
Keywords
Complete proteome
Methyltransferase
Reference proteome
RNA-binding
S-adenosyl-L-methionine
Transferase
tRNA processing
tRNA-binding
Feature
chain Probable tRNA (guanine(26)-N(2))-dimethyltransferase
Uniprot
H9JCC6
A0A2W1BJJ1
S4NU39
A0A2A4JVD7
A0A2H1V4U5
A0A212FCN3
+ More
A0A3S2LAJ8 A0A0N1PHA6 A0A2J7QEC1 A0A2J7QEA8 A0A182P4Q4 A0A182K0H0 A0A182V8A6 A0A1S4H1Y3 A0A182G3N0 A0A182XN82 A0A2C9GR22 A0A182LZX0 A0A182TK31 Q16T94 A0A1Q3EXP2 A0A182QVX7 B4P214 B0WIP5 A0A182WGI4 A0A182NBQ8 A0A1Q3EY09 Q9VK89 Q7PM72 B4IE89 B4Q3T1 A0A182Y0N0 A0A1W4VG93 A0A182UGM8 A0A1I8MVG1 A0A2M4AH45 A0A182R3T0 A0A0L0C5T6 A0A0T6B1C2 W5JES7 A0A2M4AGU6 N6TS59 U4U7Z1 B3N3U7 A0A2M4AH49 A0A1W4XGL0 A0A2M3Z7J1 A0A2M3Z7A9 A0A182F0X8 A0A2M4BID4 A0A2M4BIF2 A0A2M4BID2 A0A2M4BJ34 B4G8T1 A0A1I8NLQ9 Q29MS6 A0A1A9ZK67 A0A1B6CIS3 A0A3B0JE73 A0A084WU36 A0A182JDC6 A0A1Y1KYG8 B3MNU2 A0A1A9UH82 B4MVG1 B4JCQ3 A0A1A9WZQ4 D6WW07 B4KJ40 A0A182T004 A0A1B0AP44 A0A1A9XW12 E2BTE9 W8B9H9 A0A195FY65 F4WLB2 A0A195CRX9 E9J721 A0A158NXB7 A0A0C9QSM6 A0A151WRC9 A0A1B0FAT7 A0A1L8DRG6 A0A034WK11 A0A0J7KUH1 A0A0M3QU48 A0A0K8U6Q6 A0A1B0CYT1 A0A2A3EI55 A0A088AGI4 A0A336K6H1 A0A0A1X631 A0A151IT40 A0A0L7QVK7 A0A195BPE7 A0A1B6IAC8 A0A1B6IPS7 A0A232EPC0 E2AB22 A0A146MC45
A0A3S2LAJ8 A0A0N1PHA6 A0A2J7QEC1 A0A2J7QEA8 A0A182P4Q4 A0A182K0H0 A0A182V8A6 A0A1S4H1Y3 A0A182G3N0 A0A182XN82 A0A2C9GR22 A0A182LZX0 A0A182TK31 Q16T94 A0A1Q3EXP2 A0A182QVX7 B4P214 B0WIP5 A0A182WGI4 A0A182NBQ8 A0A1Q3EY09 Q9VK89 Q7PM72 B4IE89 B4Q3T1 A0A182Y0N0 A0A1W4VG93 A0A182UGM8 A0A1I8MVG1 A0A2M4AH45 A0A182R3T0 A0A0L0C5T6 A0A0T6B1C2 W5JES7 A0A2M4AGU6 N6TS59 U4U7Z1 B3N3U7 A0A2M4AH49 A0A1W4XGL0 A0A2M3Z7J1 A0A2M3Z7A9 A0A182F0X8 A0A2M4BID4 A0A2M4BIF2 A0A2M4BID2 A0A2M4BJ34 B4G8T1 A0A1I8NLQ9 Q29MS6 A0A1A9ZK67 A0A1B6CIS3 A0A3B0JE73 A0A084WU36 A0A182JDC6 A0A1Y1KYG8 B3MNU2 A0A1A9UH82 B4MVG1 B4JCQ3 A0A1A9WZQ4 D6WW07 B4KJ40 A0A182T004 A0A1B0AP44 A0A1A9XW12 E2BTE9 W8B9H9 A0A195FY65 F4WLB2 A0A195CRX9 E9J721 A0A158NXB7 A0A0C9QSM6 A0A151WRC9 A0A1B0FAT7 A0A1L8DRG6 A0A034WK11 A0A0J7KUH1 A0A0M3QU48 A0A0K8U6Q6 A0A1B0CYT1 A0A2A3EI55 A0A088AGI4 A0A336K6H1 A0A0A1X631 A0A151IT40 A0A0L7QVK7 A0A195BPE7 A0A1B6IAC8 A0A1B6IPS7 A0A232EPC0 E2AB22 A0A146MC45
EC Number
2.1.1.216
Pubmed
19121390
28756777
23622113
22118469
26354079
12364791
+ More
26483478 17510324 17994087 17550304 10731132 12537572 12537569 22936249 25244985 25315136 26108605 20920257 23761445 23537049 15632085 24438588 28004739 18362917 19820115 20798317 24495485 21719571 21282665 21347285 25348373 25830018 28648823 26823975
26483478 17510324 17994087 17550304 10731132 12537572 12537569 22936249 25244985 25315136 26108605 20920257 23761445 23537049 15632085 24438588 28004739 18362917 19820115 20798317 24495485 21719571 21282665 21347285 25348373 25830018 28648823 26823975
EMBL
BABH01031550
KZ150005
PZC75242.1
GAIX01011961
JAA80599.1
NWSH01000508
+ More
PCG75971.1 ODYU01000681 SOQ35870.1 AGBW02009157 OWR51499.1 RSAL01000317 RVE42592.1 KQ460882 KPJ11489.1 NEVH01015318 PNF26917.1 PNF26916.1 AAAB01008980 JXUM01141868 KQ569321 KXJ68666.1 APCN01001775 AXCM01001134 CH477657 EAT37696.1 GFDL01014966 JAV20079.1 AXCN02000684 CM000157 EDW88185.1 DS231951 EDS28622.1 GFDL01014879 JAV20166.1 AE014134 AY051836 EAA14208.5 CH480831 EDW45916.1 CM000361 CM002910 EDX04806.1 KMY89918.1 GGFK01006794 MBW40115.1 JRES01000869 KNC27650.1 LJIG01016428 KRT80631.1 ADMH02001427 ETN62576.1 GGFK01006685 MBW40006.1 APGK01056813 KB741277 ENN71141.1 KB631786 ERL86065.1 CH954177 EDV58799.1 GGFK01006795 MBW40116.1 GGFM01003742 MBW24493.1 GGFM01003662 MBW24413.1 GGFJ01003678 MBW52819.1 GGFJ01003676 MBW52817.1 GGFJ01003675 MBW52816.1 GGFJ01003677 MBW52818.1 CH479180 EDW28761.1 CH379060 EAL33617.1 GEDC01023934 JAS13364.1 OUUW01000004 SPP78953.1 ATLV01026995 KE525421 KFB53730.1 GEZM01073277 JAV65210.1 CH902620 EDV32129.1 CH963857 EDW76506.1 CH916368 EDW04217.1 KQ971361 EFA08644.2 CH933807 EDW11402.1 JXJN01001139 GL450402 EFN81030.1 GAMC01008635 JAB97920.1 KQ981169 KYN45376.1 GL888207 EGI65075.1 KQ977349 KYN03395.1 GL768421 EFZ11336.1 ADTU01029642 GBYB01006739 JAG76506.1 KQ982807 KYQ50363.1 CCAG010023607 GFDF01005006 JAV09078.1 GAKP01004285 JAC54667.1 LBMM01003110 KMQ93951.1 CP012523 ALC40043.1 GDHF01029972 JAI22342.1 AJVK01009332 KZ288235 PBC31397.1 UFQS01000093 UFQT01000093 SSW99228.1 SSX19608.1 GBXI01008154 JAD06138.1 KQ981047 KYN10034.1 KQ414726 KOC62650.1 KQ976433 KYM87327.1 GECU01023810 JAS83896.1 GECU01018787 GECU01005734 JAS88919.1 JAT01973.1 NNAY01002952 OXU20229.1 GL438234 EFN69298.1 GDHC01010912 GDHC01001281 JAQ07717.1 JAQ17348.1
PCG75971.1 ODYU01000681 SOQ35870.1 AGBW02009157 OWR51499.1 RSAL01000317 RVE42592.1 KQ460882 KPJ11489.1 NEVH01015318 PNF26917.1 PNF26916.1 AAAB01008980 JXUM01141868 KQ569321 KXJ68666.1 APCN01001775 AXCM01001134 CH477657 EAT37696.1 GFDL01014966 JAV20079.1 AXCN02000684 CM000157 EDW88185.1 DS231951 EDS28622.1 GFDL01014879 JAV20166.1 AE014134 AY051836 EAA14208.5 CH480831 EDW45916.1 CM000361 CM002910 EDX04806.1 KMY89918.1 GGFK01006794 MBW40115.1 JRES01000869 KNC27650.1 LJIG01016428 KRT80631.1 ADMH02001427 ETN62576.1 GGFK01006685 MBW40006.1 APGK01056813 KB741277 ENN71141.1 KB631786 ERL86065.1 CH954177 EDV58799.1 GGFK01006795 MBW40116.1 GGFM01003742 MBW24493.1 GGFM01003662 MBW24413.1 GGFJ01003678 MBW52819.1 GGFJ01003676 MBW52817.1 GGFJ01003675 MBW52816.1 GGFJ01003677 MBW52818.1 CH479180 EDW28761.1 CH379060 EAL33617.1 GEDC01023934 JAS13364.1 OUUW01000004 SPP78953.1 ATLV01026995 KE525421 KFB53730.1 GEZM01073277 JAV65210.1 CH902620 EDV32129.1 CH963857 EDW76506.1 CH916368 EDW04217.1 KQ971361 EFA08644.2 CH933807 EDW11402.1 JXJN01001139 GL450402 EFN81030.1 GAMC01008635 JAB97920.1 KQ981169 KYN45376.1 GL888207 EGI65075.1 KQ977349 KYN03395.1 GL768421 EFZ11336.1 ADTU01029642 GBYB01006739 JAG76506.1 KQ982807 KYQ50363.1 CCAG010023607 GFDF01005006 JAV09078.1 GAKP01004285 JAC54667.1 LBMM01003110 KMQ93951.1 CP012523 ALC40043.1 GDHF01029972 JAI22342.1 AJVK01009332 KZ288235 PBC31397.1 UFQS01000093 UFQT01000093 SSW99228.1 SSX19608.1 GBXI01008154 JAD06138.1 KQ981047 KYN10034.1 KQ414726 KOC62650.1 KQ976433 KYM87327.1 GECU01023810 JAS83896.1 GECU01018787 GECU01005734 JAS88919.1 JAT01973.1 NNAY01002952 OXU20229.1 GL438234 EFN69298.1 GDHC01010912 GDHC01001281 JAQ07717.1 JAQ17348.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000235965
+ More
UP000075885 UP000075881 UP000075903 UP000069940 UP000249989 UP000076407 UP000075840 UP000075883 UP000075902 UP000008820 UP000075886 UP000002282 UP000002320 UP000075920 UP000075884 UP000000803 UP000007062 UP000001292 UP000000304 UP000076408 UP000192221 UP000095301 UP000075900 UP000037069 UP000000673 UP000019118 UP000030742 UP000008711 UP000192223 UP000069272 UP000008744 UP000095300 UP000001819 UP000092445 UP000268350 UP000030765 UP000075880 UP000007801 UP000078200 UP000007798 UP000001070 UP000091820 UP000007266 UP000009192 UP000075901 UP000092460 UP000092443 UP000008237 UP000078541 UP000007755 UP000078542 UP000005205 UP000075809 UP000092444 UP000036403 UP000092553 UP000092462 UP000242457 UP000005203 UP000078492 UP000053825 UP000078540 UP000215335 UP000000311
UP000075885 UP000075881 UP000075903 UP000069940 UP000249989 UP000076407 UP000075840 UP000075883 UP000075902 UP000008820 UP000075886 UP000002282 UP000002320 UP000075920 UP000075884 UP000000803 UP000007062 UP000001292 UP000000304 UP000076408 UP000192221 UP000095301 UP000075900 UP000037069 UP000000673 UP000019118 UP000030742 UP000008711 UP000192223 UP000069272 UP000008744 UP000095300 UP000001819 UP000092445 UP000268350 UP000030765 UP000075880 UP000007801 UP000078200 UP000007798 UP000001070 UP000091820 UP000007266 UP000009192 UP000075901 UP000092460 UP000092443 UP000008237 UP000078541 UP000007755 UP000078542 UP000005205 UP000075809 UP000092444 UP000036403 UP000092553 UP000092462 UP000242457 UP000005203 UP000078492 UP000053825 UP000078540 UP000215335 UP000000311
Interpro
IPR029063
SAM-dependent_MTases
+ More
IPR002905 Trm1
IPR042296 tRNA_met_Trm1_C
IPR029382 NCU-G1
IPR016024 ARM-type_fold
IPR034085 TOG
IPR038737 SF3b_su1-like
IPR022179 DUF3695
IPR011989 ARM-like
IPR015016 SF3b_su1
IPR036812 NADP_OxRdtase_dom_sf
IPR018170 Aldo/ket_reductase_CS
IPR020471 Aldo/keto_reductase
IPR023210 NADP_OxRdtase_dom
IPR002905 Trm1
IPR042296 tRNA_met_Trm1_C
IPR029382 NCU-G1
IPR016024 ARM-type_fold
IPR034085 TOG
IPR038737 SF3b_su1-like
IPR022179 DUF3695
IPR011989 ARM-like
IPR015016 SF3b_su1
IPR036812 NADP_OxRdtase_dom_sf
IPR018170 Aldo/ket_reductase_CS
IPR020471 Aldo/keto_reductase
IPR023210 NADP_OxRdtase_dom
Gene 3D
ProteinModelPortal
H9JCC6
A0A2W1BJJ1
S4NU39
A0A2A4JVD7
A0A2H1V4U5
A0A212FCN3
+ More
A0A3S2LAJ8 A0A0N1PHA6 A0A2J7QEC1 A0A2J7QEA8 A0A182P4Q4 A0A182K0H0 A0A182V8A6 A0A1S4H1Y3 A0A182G3N0 A0A182XN82 A0A2C9GR22 A0A182LZX0 A0A182TK31 Q16T94 A0A1Q3EXP2 A0A182QVX7 B4P214 B0WIP5 A0A182WGI4 A0A182NBQ8 A0A1Q3EY09 Q9VK89 Q7PM72 B4IE89 B4Q3T1 A0A182Y0N0 A0A1W4VG93 A0A182UGM8 A0A1I8MVG1 A0A2M4AH45 A0A182R3T0 A0A0L0C5T6 A0A0T6B1C2 W5JES7 A0A2M4AGU6 N6TS59 U4U7Z1 B3N3U7 A0A2M4AH49 A0A1W4XGL0 A0A2M3Z7J1 A0A2M3Z7A9 A0A182F0X8 A0A2M4BID4 A0A2M4BIF2 A0A2M4BID2 A0A2M4BJ34 B4G8T1 A0A1I8NLQ9 Q29MS6 A0A1A9ZK67 A0A1B6CIS3 A0A3B0JE73 A0A084WU36 A0A182JDC6 A0A1Y1KYG8 B3MNU2 A0A1A9UH82 B4MVG1 B4JCQ3 A0A1A9WZQ4 D6WW07 B4KJ40 A0A182T004 A0A1B0AP44 A0A1A9XW12 E2BTE9 W8B9H9 A0A195FY65 F4WLB2 A0A195CRX9 E9J721 A0A158NXB7 A0A0C9QSM6 A0A151WRC9 A0A1B0FAT7 A0A1L8DRG6 A0A034WK11 A0A0J7KUH1 A0A0M3QU48 A0A0K8U6Q6 A0A1B0CYT1 A0A2A3EI55 A0A088AGI4 A0A336K6H1 A0A0A1X631 A0A151IT40 A0A0L7QVK7 A0A195BPE7 A0A1B6IAC8 A0A1B6IPS7 A0A232EPC0 E2AB22 A0A146MC45
A0A3S2LAJ8 A0A0N1PHA6 A0A2J7QEC1 A0A2J7QEA8 A0A182P4Q4 A0A182K0H0 A0A182V8A6 A0A1S4H1Y3 A0A182G3N0 A0A182XN82 A0A2C9GR22 A0A182LZX0 A0A182TK31 Q16T94 A0A1Q3EXP2 A0A182QVX7 B4P214 B0WIP5 A0A182WGI4 A0A182NBQ8 A0A1Q3EY09 Q9VK89 Q7PM72 B4IE89 B4Q3T1 A0A182Y0N0 A0A1W4VG93 A0A182UGM8 A0A1I8MVG1 A0A2M4AH45 A0A182R3T0 A0A0L0C5T6 A0A0T6B1C2 W5JES7 A0A2M4AGU6 N6TS59 U4U7Z1 B3N3U7 A0A2M4AH49 A0A1W4XGL0 A0A2M3Z7J1 A0A2M3Z7A9 A0A182F0X8 A0A2M4BID4 A0A2M4BIF2 A0A2M4BID2 A0A2M4BJ34 B4G8T1 A0A1I8NLQ9 Q29MS6 A0A1A9ZK67 A0A1B6CIS3 A0A3B0JE73 A0A084WU36 A0A182JDC6 A0A1Y1KYG8 B3MNU2 A0A1A9UH82 B4MVG1 B4JCQ3 A0A1A9WZQ4 D6WW07 B4KJ40 A0A182T004 A0A1B0AP44 A0A1A9XW12 E2BTE9 W8B9H9 A0A195FY65 F4WLB2 A0A195CRX9 E9J721 A0A158NXB7 A0A0C9QSM6 A0A151WRC9 A0A1B0FAT7 A0A1L8DRG6 A0A034WK11 A0A0J7KUH1 A0A0M3QU48 A0A0K8U6Q6 A0A1B0CYT1 A0A2A3EI55 A0A088AGI4 A0A336K6H1 A0A0A1X631 A0A151IT40 A0A0L7QVK7 A0A195BPE7 A0A1B6IAC8 A0A1B6IPS7 A0A232EPC0 E2AB22 A0A146MC45
PDB
2YTZ
E-value=7.47797e-47,
Score=473
Ontologies
GO
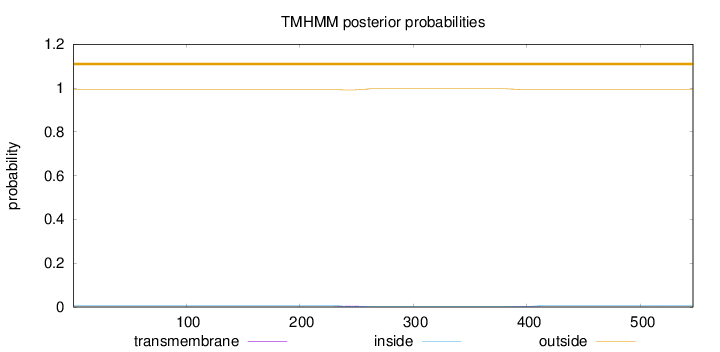
Topology
Length:
546
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17474
Exp number, first 60 AAs:
0.00028
Total prob of N-in:
0.00779
outside
1 - 546
Population Genetic Test Statistics
Pi
19.095209
Theta
19.58061
Tajima's D
0.027234
CLR
1.284905
CSRT
0.381480925953702
Interpretation
Uncertain
