Gene
KWMTBOMO12737
Pre Gene Modal
BGIBMGA007193
Annotation
PREDICTED:_m7GpppN-mRNA_hydrolase-like_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 4.069
Sequence
CDS
ATGAATTCAAGTCATAACAAACACTCTATTCCAATAGATATATTAGATGACCTTTGTAGTCGTTTCATAATAAATCTACCAGCAGAAGATAGAGGAAACCTAGTGAGGATATGCTTCCAGATCGAATTGGCTCATTGGTTTTATTTGGATTATTACTGCACTGACGAATCGAAGAAAGTATACCCCTGTGGTATTAGAGAGTTTGCAGCACATATTTTCCAACATGTCCCGCAACTCCGAGAACATGTTAGCAGTCTAGATGCAGTTTTAGATAACTGGAGAGAATATAAACAGACTGTACCTACATACGGAGCTATACTGCTCGATGATGACCTCTCTCATGTGCTTCTTGTACAATCATATTGGACAAAAGCCTCCTGGGGTTTCCCTAAAGGGAAGGTCAATGAAGATGAGGAGCCCTGGAAGTGTGCTACTAGAGAGGTTCTAGAAGAGACAGGTTTTGATATAAGCAATTTAATCAATAAAAACGATTACATAGAAGCCGTAACACACGACCAAATCGCTCGTTTGTACATTATTGGAAATATTCCCAGAGACACAAAGTTCCAACCTCGCACACGAAACGAGATCAAAGCGTGCGAATGGTTCCCATTGGCGGATTTACCGGCCAATAAAAAAGACATGACGCCCAAAGTCAAAATGGGGGTTAGCCCTAATGCGTTCTTCATGGTCCTACCTTTCGTCAAACGAATGAGACGATGGGTTGCCGAACGCAGCTCCAAGGTATTCACGAACAGTCGTCGAACTCGTCACAAATCGATGGGAGATTTAGAAGCTTCAACGAGCCAGAATAAAAACAAGACCATATCGCAGGGACTGCAAAACGAGATAAATGAATACCAACAAAATTCTGGTCACAAAAATGATTATCAGAATCATGGAAGCAATACAAAAAATGTTAATGGTAATAAAAATGGTGGCAATGGTAACAAAAATGGTGCTAATGGCAATAATAGTGCAAGGAAGGAAAAGAAGGCTGCTAAAAGACAGTTGTTCACGCCACAGAACATCCAGTCGAATAATATTTCACCGGTACAAAAGGAAGACAGTGTTGCCGAGGCTAAAATAGATGAAAGCAAATTAAGCAATTTCATTGCTCCATCTTGGGCAGACTTCAAGTTTGACAAAAGGGCAATTCTTGATTGTCTCACTTGA
Protein
MNSSHNKHSIPIDILDDLCSRFIINLPAEDRGNLVRICFQIELAHWFYLDYYCTDESKKVYPCGIREFAAHIFQHVPQLREHVSSLDAVLDNWREYKQTVPTYGAILLDDDLSHVLLVQSYWTKASWGFPKGKVNEDEEPWKCATREVLEETGFDISNLINKNDYIEAVTHDQIARLYIIGNIPRDTKFQPRTRNEIKACEWFPLADLPANKKDMTPKVKMGVSPNAFFMVLPFVKRMRRWVAERSSKVFTNSRRTRHKSMGDLEASTSQNKNKTISQGLQNEINEYQQNSGHKNDYQNHGSNTKNVNGNKNGGNGNKNGANGNNSARKEKKAAKRQLFTPQNIQSNNISPVQKEDSVAEAKIDESKLSNFIAPSWADFKFDKRAILDCLT
Summary
Similarity
Belongs to the serpin family.
Uniprot
H9JCE8
A0A437AWP4
A0A2A4JVE7
A0A194QG70
A0A212FCQ5
A0A2H1VJX0
+ More
S4PHZ0 A0A2W1BPJ5 A0A0L7KU06 A0A0N1IG90 A0A2J7QZ12 A0A2P8YIN6 A0A1B6E4F0 A0A067QES9 E2BP76 A0A0L7RHG1 A0A1B6I4Q5 A0A088ACQ5 E2AD15 A0A232EG53 F4W990 A0A1B6MNL8 A0A195B513 A0A158P0V8 A0A195EEL7 A0A151WKJ5 A0A195F3C1 A0A0M8ZSC5 K7IMG9 D6WCP2 A0A0T6B9Z0 A0A1W4X3S3 A0A310SSN5 A0A1Y1KPC7 A0A1E1XPV5 A0A2R5LL72 A0A293MZF2 E9G2H3 A0A1B0GK27 A0A0P5DV70 A0A0K8TS82 A0A1L8DFX5 A0A0P6E3J7 A0A1B0GN89 A0A1A9WRB0 A0A0K8VM28 A0A0A1WZU8 A0A0A1X8P8 Q2M0K0 B4HIK0 W8CAE2 A0A0J9RVT7 A0A0J9RX44 A0A0R1E8B2 B4IU38 A0A2L2Y0K4 Q5U127 Q9VUU4 A0A182QET0 A0A1W4VAY2 A0A1W4VP05 A0A1S3IZY7 A0A0Q5UK15 A0A0P6I8W9 B3M9V9 B4MXW4 Q1DGJ5
S4PHZ0 A0A2W1BPJ5 A0A0L7KU06 A0A0N1IG90 A0A2J7QZ12 A0A2P8YIN6 A0A1B6E4F0 A0A067QES9 E2BP76 A0A0L7RHG1 A0A1B6I4Q5 A0A088ACQ5 E2AD15 A0A232EG53 F4W990 A0A1B6MNL8 A0A195B513 A0A158P0V8 A0A195EEL7 A0A151WKJ5 A0A195F3C1 A0A0M8ZSC5 K7IMG9 D6WCP2 A0A0T6B9Z0 A0A1W4X3S3 A0A310SSN5 A0A1Y1KPC7 A0A1E1XPV5 A0A2R5LL72 A0A293MZF2 E9G2H3 A0A1B0GK27 A0A0P5DV70 A0A0K8TS82 A0A1L8DFX5 A0A0P6E3J7 A0A1B0GN89 A0A1A9WRB0 A0A0K8VM28 A0A0A1WZU8 A0A0A1X8P8 Q2M0K0 B4HIK0 W8CAE2 A0A0J9RVT7 A0A0J9RX44 A0A0R1E8B2 B4IU38 A0A2L2Y0K4 Q5U127 Q9VUU4 A0A182QET0 A0A1W4VAY2 A0A1W4VP05 A0A1S3IZY7 A0A0Q5UK15 A0A0P6I8W9 B3M9V9 B4MXW4 Q1DGJ5
Pubmed
19121390
23643815
26354079
22118469
23622113
28756777
+ More
26227816 29403074 24845553 20798317 28648823 21719571 21347285 20075255 18362917 19820115 28004739 29209593 21292972 26369729 25830018 15632085 17994087 24495485 22936249 17550304 26561354 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324
26227816 29403074 24845553 20798317 28648823 21719571 21347285 20075255 18362917 19820115 28004739 29209593 21292972 26369729 25830018 15632085 17994087 24495485 22936249 17550304 26561354 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324
EMBL
BABH01031549
JX027620
AGE13483.1
RSAL01000317
RVE42594.1
NWSH01000508
+ More
PCG75981.1 KQ458981 KPJ04487.1 AGBW02009157 OWR51497.1 ODYU01002960 SOQ41123.1 GAIX01005620 JAA86940.1 KZ150005 PZC75237.1 JTDY01005870 KOB66531.1 KQ460882 KPJ11491.1 NEVH01009080 PNF33817.1 PYGN01000568 PSN44115.1 GEDC01004514 JAS32784.1 KK853714 KDQ94595.1 GL449553 EFN82496.1 KQ414590 KOC70407.1 GECU01025826 JAS81880.1 GL438658 EFN68651.1 NNAY01004881 OXU17302.1 GL888002 EGI69272.1 GEBQ01002437 JAT37540.1 KQ976598 KYM79586.1 ADTU01001034 KQ979039 KYN23252.1 KQ983002 KYQ48403.1 KQ981855 KYN34881.1 KQ435876 KOX70145.1 KQ971317 EEZ97803.1 LJIG01002862 KRT84078.1 KQ760478 OAD60243.1 GEZM01081834 JAV61515.1 GFAA01002106 JAU01329.1 GGLE01006150 MBY10276.1 GFWV01021455 MAA46183.1 GL732530 EFX86250.1 AJWK01022514 GDIP01151103 JAJ72299.1 GDAI01000813 JAI16790.1 GFDF01008738 JAV05346.1 GDIQ01069518 JAN25219.1 AJVK01013078 GDHF01012385 JAI39929.1 GBXI01009698 JAD04594.1 GBXI01007036 JAD07256.1 CH379069 EAL30932.3 CH480815 EDW41630.1 GAMC01005826 JAC00730.1 CM002912 KMY99831.1 KMY99829.1 CH891771 KRK05285.1 EDW99901.1 IAAA01004844 LAA00635.1 BT016065 AE014296 AAV36950.1 AFH04447.1 AAF49579.4 AXCN02001739 CH954178 KQS44128.1 GDIQ01007869 JAN86868.1 CH902618 EDV40150.1 CH963876 EDW76883.2 CH901524 EAT32274.1
PCG75981.1 KQ458981 KPJ04487.1 AGBW02009157 OWR51497.1 ODYU01002960 SOQ41123.1 GAIX01005620 JAA86940.1 KZ150005 PZC75237.1 JTDY01005870 KOB66531.1 KQ460882 KPJ11491.1 NEVH01009080 PNF33817.1 PYGN01000568 PSN44115.1 GEDC01004514 JAS32784.1 KK853714 KDQ94595.1 GL449553 EFN82496.1 KQ414590 KOC70407.1 GECU01025826 JAS81880.1 GL438658 EFN68651.1 NNAY01004881 OXU17302.1 GL888002 EGI69272.1 GEBQ01002437 JAT37540.1 KQ976598 KYM79586.1 ADTU01001034 KQ979039 KYN23252.1 KQ983002 KYQ48403.1 KQ981855 KYN34881.1 KQ435876 KOX70145.1 KQ971317 EEZ97803.1 LJIG01002862 KRT84078.1 KQ760478 OAD60243.1 GEZM01081834 JAV61515.1 GFAA01002106 JAU01329.1 GGLE01006150 MBY10276.1 GFWV01021455 MAA46183.1 GL732530 EFX86250.1 AJWK01022514 GDIP01151103 JAJ72299.1 GDAI01000813 JAI16790.1 GFDF01008738 JAV05346.1 GDIQ01069518 JAN25219.1 AJVK01013078 GDHF01012385 JAI39929.1 GBXI01009698 JAD04594.1 GBXI01007036 JAD07256.1 CH379069 EAL30932.3 CH480815 EDW41630.1 GAMC01005826 JAC00730.1 CM002912 KMY99831.1 KMY99829.1 CH891771 KRK05285.1 EDW99901.1 IAAA01004844 LAA00635.1 BT016065 AE014296 AAV36950.1 AFH04447.1 AAF49579.4 AXCN02001739 CH954178 KQS44128.1 GDIQ01007869 JAN86868.1 CH902618 EDV40150.1 CH963876 EDW76883.2 CH901524 EAT32274.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000037510
+ More
UP000053240 UP000235965 UP000245037 UP000027135 UP000008237 UP000053825 UP000005203 UP000000311 UP000215335 UP000007755 UP000078540 UP000005205 UP000078492 UP000075809 UP000078541 UP000053105 UP000002358 UP000007266 UP000192223 UP000000305 UP000092461 UP000092462 UP000091820 UP000001819 UP000001292 UP000002282 UP000000803 UP000075886 UP000192221 UP000085678 UP000008711 UP000007801 UP000007798 UP000008820
UP000053240 UP000235965 UP000245037 UP000027135 UP000008237 UP000053825 UP000005203 UP000000311 UP000215335 UP000007755 UP000078540 UP000005205 UP000078492 UP000075809 UP000078541 UP000053105 UP000002358 UP000007266 UP000192223 UP000000305 UP000092461 UP000092462 UP000091820 UP000001819 UP000001292 UP000002282 UP000000803 UP000075886 UP000192221 UP000085678 UP000008711 UP000007801 UP000007798 UP000008820
Interpro
Gene 3D
ProteinModelPortal
H9JCE8
A0A437AWP4
A0A2A4JVE7
A0A194QG70
A0A212FCQ5
A0A2H1VJX0
+ More
S4PHZ0 A0A2W1BPJ5 A0A0L7KU06 A0A0N1IG90 A0A2J7QZ12 A0A2P8YIN6 A0A1B6E4F0 A0A067QES9 E2BP76 A0A0L7RHG1 A0A1B6I4Q5 A0A088ACQ5 E2AD15 A0A232EG53 F4W990 A0A1B6MNL8 A0A195B513 A0A158P0V8 A0A195EEL7 A0A151WKJ5 A0A195F3C1 A0A0M8ZSC5 K7IMG9 D6WCP2 A0A0T6B9Z0 A0A1W4X3S3 A0A310SSN5 A0A1Y1KPC7 A0A1E1XPV5 A0A2R5LL72 A0A293MZF2 E9G2H3 A0A1B0GK27 A0A0P5DV70 A0A0K8TS82 A0A1L8DFX5 A0A0P6E3J7 A0A1B0GN89 A0A1A9WRB0 A0A0K8VM28 A0A0A1WZU8 A0A0A1X8P8 Q2M0K0 B4HIK0 W8CAE2 A0A0J9RVT7 A0A0J9RX44 A0A0R1E8B2 B4IU38 A0A2L2Y0K4 Q5U127 Q9VUU4 A0A182QET0 A0A1W4VAY2 A0A1W4VP05 A0A1S3IZY7 A0A0Q5UK15 A0A0P6I8W9 B3M9V9 B4MXW4 Q1DGJ5
S4PHZ0 A0A2W1BPJ5 A0A0L7KU06 A0A0N1IG90 A0A2J7QZ12 A0A2P8YIN6 A0A1B6E4F0 A0A067QES9 E2BP76 A0A0L7RHG1 A0A1B6I4Q5 A0A088ACQ5 E2AD15 A0A232EG53 F4W990 A0A1B6MNL8 A0A195B513 A0A158P0V8 A0A195EEL7 A0A151WKJ5 A0A195F3C1 A0A0M8ZSC5 K7IMG9 D6WCP2 A0A0T6B9Z0 A0A1W4X3S3 A0A310SSN5 A0A1Y1KPC7 A0A1E1XPV5 A0A2R5LL72 A0A293MZF2 E9G2H3 A0A1B0GK27 A0A0P5DV70 A0A0K8TS82 A0A1L8DFX5 A0A0P6E3J7 A0A1B0GN89 A0A1A9WRB0 A0A0K8VM28 A0A0A1WZU8 A0A0A1X8P8 Q2M0K0 B4HIK0 W8CAE2 A0A0J9RVT7 A0A0J9RX44 A0A0R1E8B2 B4IU38 A0A2L2Y0K4 Q5U127 Q9VUU4 A0A182QET0 A0A1W4VAY2 A0A1W4VP05 A0A1S3IZY7 A0A0Q5UK15 A0A0P6I8W9 B3M9V9 B4MXW4 Q1DGJ5
PDB
5KQ4
E-value=3.98782e-50,
Score=500
Ontologies
GO
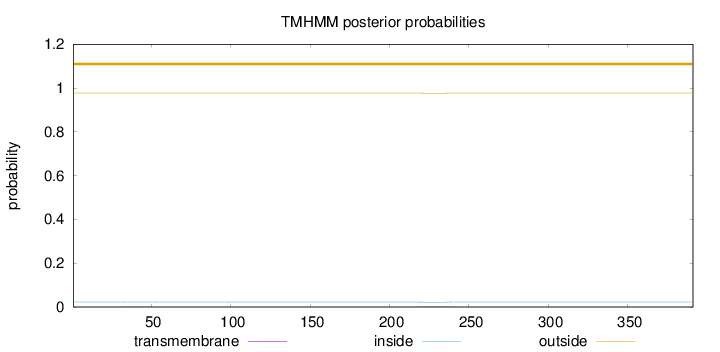
Topology
Length:
391
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03368
Exp number, first 60 AAs:
0.00649
Total prob of N-in:
0.02278
outside
1 - 391
Population Genetic Test Statistics
Pi
30.523091
Theta
23.069658
Tajima's D
1.023554
CLR
0.351875
CSRT
0.666066696665167
Interpretation
Uncertain
