Pre Gene Modal
BGIBMGA007173
Annotation
putative_Syntaxin-18_[Danaus_plexippus]
Full name
GPI inositol-deacylase
+ More
Syntaxin-18
Syntaxin-18
Location in the cell
Nuclear Reliability : 3.022
Sequence
CDS
ATGGATATAACACCATTATTTAAGGCTTGCATAAAAACTGTTAAAACTCGTAACAAGGCTTTCGGCATACACAGTCCGATTAGCGAAGATAAGCAACGCATATTGAGGGTGAAATCAAAAAATGCTTTCATGGCAACAGCTAAGGATATTTGTTCACAAATTACAAAGCTTAGGGACTTTTTATTGGAACACCGCGATAAATATTTAAGCTTCTTCAATAATGTTACTGGAGACGATATGTCTGATGAAGAGCGCGACCAAATAGACACCGGTGCGCAACGTATCATGAACACATGCTCACACCTGCTAAAAGAGTTCAGGAATGATAATCGAAGAACTACAGTGACCCCACAAACTAGGGAATATATGGACTCAGTCGTAGATTTAATTGATGCTTATTTAAAAGCTGTTTGTAAAGTGCATAGTGAATTGAAAGCAATGAGAGTTAAGCGAGCTTTAGATATGAGAAAATTGTCTAGACTTGAGCTACCAGCGACCAAATCTAGTATTCCTAATCCGTTTACACTAGATAAGGTAATAGAAAAAGAGAATGAGACAGAAGATGAAAAAGAAAATACAAAAACAAAATTAGACTTATCAGAGAATGAGATGGCAATTATGTCTGATGAAGGAGAGTTGACTTCAGAAGAGTTGCAAATGTTTGAGTCGGAGAATGTACAGCTGTTAAATGAACTAAACAGTATGACGGAAGAAGTTAAACAAATTGAAAGCAAAGTACTACACATTGCAGAACTTCAAGAAATATTTACTGAAAAGGTTTTACAACAAGAACAAGACATAGATAGAATATCGAATACAGTTGTCGGGGCAACTGAAAACGTTAAGGATGCCAATGAACAGATTAAGCAAGCTATACAAAGAAACGCAGGCCTAAGAGTATATGTTCTGTTCTTCTTACTGGTCATGTCGTTCAGTTTGCTGTTCCTAGATTGGTATAATGATTAA
Protein
MDITPLFKACIKTVKTRNKAFGIHSPISEDKQRILRVKSKNAFMATAKDICSQITKLRDFLLEHRDKYLSFFNNVTGDDMSDEERDQIDTGAQRIMNTCSHLLKEFRNDNRRTTVTPQTREYMDSVVDLIDAYLKAVCKVHSELKAMRVKRALDMRKLSRLELPATKSSIPNPFTLDKVIEKENETEDEKENTKTKLDLSENEMAIMSDEGELTSEELQMFESENVQLLNELNSMTEEVKQIESKVLHIAELQEIFTEKVLQQEQDIDRISNTVVGATENVKDANEQIKQAIQRNAGLRVYVLFFLLVMSFSLLFLDWYND
Summary
Description
Involved in inositol deacylation of GPI-anchored proteins which plays important roles in the quality control and ER-associated degradation of GPI-anchored proteins.
Syntaxin that may be involved in targeting and fusion of Golgi-derived retrograde transport vesicles with the ER.
Syntaxin that may be involved in targeting and fusion of Golgi-derived retrograde transport vesicles with the ER.
Similarity
Belongs to the GPI inositol-deacylase family.
Belongs to the syntaxin family.
Belongs to the syntaxin family.
Keywords
Coiled coil
Complete proteome
Endoplasmic reticulum
ER-Golgi transport
Membrane
Protein transport
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Syntaxin-18
Uniprot
H9JCC8
S4NY43
A0A1E1VYH1
A0A0N1IER1
A0A194QG14
A0A2W1BK28
+ More
A0A2A4JX64 A0A3S2P5X2 A0A2H1VLK1 A0A212FCM4 A0A0L7KTD5 A0A2P8YJH0 A0A2J7R1B9 A0A067RBV0 A0A1B6CVB8 A0A1B6IXZ2 A0A1B6LMT0 A0A0M9A980 A0A158NM72 A0A195BWY4 A0A151JLI5 A0A151X1X5 F4WK14 A0A151IE89 A0A151JTN8 A0A2A3ENI4 A0A1B6GGX0 A0A088A105 A0A0L7QUJ3 E9J4J2 A0A154PSE5 A0A0J7KWA0 A0A2D0QAZ0 E2C2S7 A0A3B4C8D5 E3TD31 E2ABW1 A0A3P8S1C0 A0A3Q4HLZ3 A0A3P8PF05 A0A3P9DNQ3 A0A3B4FAU8 A0A3Q2Q9M6 A0A3Q2W676 A0A3Q1JUH6 A0A3B5B3E2 A0A3Q3GZQ0 A0A147A0T3 A0A3B4Y9S8 N6U9N4 A0A0F8ACM9 E6ZG47 A0A091I341 A0A3Q1C9B9 A0A401NYE2 G3P074 A0A3B4VMC3 A0A0S7HVT3 A0A1S3FTR9 A0A3B5RFN9 A0A087XLM6 A0A3B3XXJ9 A0A3B3U4K1 A0A3Q3MH21 E0VJI0 A0A1Y1L5E4 V9KWT3 A0A401SAA2 H2MQE0 A0A3Q1G422 A0A3L8DKV4 A0A026WGF9 A0A3P9KSA1 A0A3Q2ZAQ7 A0A3P9MVI9 A0A2U9BGI8 I3KTK6 A0A3Q2DPZ2 A0A3S2MCD1 A0A1S3L997 A0A3P9HFS6 A0A3B4VMC8 A0A218VCW3 A0A2I4BBN6 E1C1M8 F6R8M8 A0A151P9V2 W5MV18 A0A452IR59 A0A2K5ID15 A0A3B5JZ15 A0A1W4WHH3 A0A1A8IQP9 Q4VBI7 A0A1A8C6F6 A0A1A7ZQV6 A0A1A8EYI7 A0A023F083 A0A3B3RJ61 H3D095
A0A2A4JX64 A0A3S2P5X2 A0A2H1VLK1 A0A212FCM4 A0A0L7KTD5 A0A2P8YJH0 A0A2J7R1B9 A0A067RBV0 A0A1B6CVB8 A0A1B6IXZ2 A0A1B6LMT0 A0A0M9A980 A0A158NM72 A0A195BWY4 A0A151JLI5 A0A151X1X5 F4WK14 A0A151IE89 A0A151JTN8 A0A2A3ENI4 A0A1B6GGX0 A0A088A105 A0A0L7QUJ3 E9J4J2 A0A154PSE5 A0A0J7KWA0 A0A2D0QAZ0 E2C2S7 A0A3B4C8D5 E3TD31 E2ABW1 A0A3P8S1C0 A0A3Q4HLZ3 A0A3P8PF05 A0A3P9DNQ3 A0A3B4FAU8 A0A3Q2Q9M6 A0A3Q2W676 A0A3Q1JUH6 A0A3B5B3E2 A0A3Q3GZQ0 A0A147A0T3 A0A3B4Y9S8 N6U9N4 A0A0F8ACM9 E6ZG47 A0A091I341 A0A3Q1C9B9 A0A401NYE2 G3P074 A0A3B4VMC3 A0A0S7HVT3 A0A1S3FTR9 A0A3B5RFN9 A0A087XLM6 A0A3B3XXJ9 A0A3B3U4K1 A0A3Q3MH21 E0VJI0 A0A1Y1L5E4 V9KWT3 A0A401SAA2 H2MQE0 A0A3Q1G422 A0A3L8DKV4 A0A026WGF9 A0A3P9KSA1 A0A3Q2ZAQ7 A0A3P9MVI9 A0A2U9BGI8 I3KTK6 A0A3Q2DPZ2 A0A3S2MCD1 A0A1S3L997 A0A3P9HFS6 A0A3B4VMC8 A0A218VCW3 A0A2I4BBN6 E1C1M8 F6R8M8 A0A151P9V2 W5MV18 A0A452IR59 A0A2K5ID15 A0A3B5JZ15 A0A1W4WHH3 A0A1A8IQP9 Q4VBI7 A0A1A8C6F6 A0A1A7ZQV6 A0A1A8EYI7 A0A023F083 A0A3B3RJ61 H3D095
EC Number
3.1.-.-
Pubmed
EMBL
BABH01031549
GAIX01013945
JAA78615.1
GDQN01011333
JAT79721.1
KQ460882
+ More
KPJ11492.1 KQ458981 KPJ04488.1 KZ150005 PZC75238.1 NWSH01000508 PCG75982.1 RSAL01000317 RVE42595.1 ODYU01002960 SOQ41124.1 AGBW02009157 OWR51496.1 JTDY01005870 KOB66532.1 PYGN01000551 PSN44400.1 NEVH01008208 PNF34624.1 PNF34626.1 KK852804 KDR16158.1 GEDC01020125 GEDC01017041 GEDC01016849 GEDC01003051 JAS17173.1 JAS20257.1 JAS20449.1 JAS34247.1 GECU01015899 JAS91807.1 GEBQ01014926 JAT25051.1 KQ435716 KOX79066.1 ADTU01020331 KQ976395 KYM93092.1 KQ979011 KYN26625.1 KQ982584 KYQ54405.1 GL888190 EGI65464.1 KQ977890 KYM98985.1 KQ981873 KYN34024.1 KZ288215 PBC32601.1 GECZ01008093 JAS61676.1 KQ414735 KOC62285.1 GL768105 EFZ12275.1 KQ435127 KZC14793.1 LBMM01002638 KMQ94539.1 GL452203 EFN77762.1 GU588261 ADO28217.1 GL438386 EFN69056.1 GCES01014498 JAR71825.1 APGK01043560 KB741015 KB632161 ENN75317.1 ERL89245.1 KQ042685 KKF11541.1 FQ310506 CBN81147.1 KL218144 KFP02637.1 BFAA01002888 GCB65889.1 GBYX01436381 JAO44959.1 AYCK01005639 AYCK01005640 DS235222 EEB13536.1 GEZM01066267 JAV68028.1 JW870607 AFP03125.1 BEZZ01000161 GCC27337.1 QOIP01000007 RLU20539.1 KK107238 EZA54781.1 CP026248 AWP03155.1 AERX01006897 CM012441 RVE72570.1 MUZQ01000010 OWK63688.1 AADN05000017 AAMC01088192 AAMC01088193 AAMC01088194 AAMC01088195 AAMC01088196 AAMC01088197 AAMC01088198 AAMC01088199 AAMC01088200 AKHW03000533 KYO45750.1 AHAT01001185 AHAT01001186 AHAT01001187 HAED01013205 HAEE01008207 SBQ99650.1 BC095763 AAH95763.1 HADZ01010707 HAEA01016296 SBP74648.1 HADY01006206 HAEJ01015231 SBP44691.1 HAEB01004392 SBQ50919.1 GBBI01003807 JAC14905.1
KPJ11492.1 KQ458981 KPJ04488.1 KZ150005 PZC75238.1 NWSH01000508 PCG75982.1 RSAL01000317 RVE42595.1 ODYU01002960 SOQ41124.1 AGBW02009157 OWR51496.1 JTDY01005870 KOB66532.1 PYGN01000551 PSN44400.1 NEVH01008208 PNF34624.1 PNF34626.1 KK852804 KDR16158.1 GEDC01020125 GEDC01017041 GEDC01016849 GEDC01003051 JAS17173.1 JAS20257.1 JAS20449.1 JAS34247.1 GECU01015899 JAS91807.1 GEBQ01014926 JAT25051.1 KQ435716 KOX79066.1 ADTU01020331 KQ976395 KYM93092.1 KQ979011 KYN26625.1 KQ982584 KYQ54405.1 GL888190 EGI65464.1 KQ977890 KYM98985.1 KQ981873 KYN34024.1 KZ288215 PBC32601.1 GECZ01008093 JAS61676.1 KQ414735 KOC62285.1 GL768105 EFZ12275.1 KQ435127 KZC14793.1 LBMM01002638 KMQ94539.1 GL452203 EFN77762.1 GU588261 ADO28217.1 GL438386 EFN69056.1 GCES01014498 JAR71825.1 APGK01043560 KB741015 KB632161 ENN75317.1 ERL89245.1 KQ042685 KKF11541.1 FQ310506 CBN81147.1 KL218144 KFP02637.1 BFAA01002888 GCB65889.1 GBYX01436381 JAO44959.1 AYCK01005639 AYCK01005640 DS235222 EEB13536.1 GEZM01066267 JAV68028.1 JW870607 AFP03125.1 BEZZ01000161 GCC27337.1 QOIP01000007 RLU20539.1 KK107238 EZA54781.1 CP026248 AWP03155.1 AERX01006897 CM012441 RVE72570.1 MUZQ01000010 OWK63688.1 AADN05000017 AAMC01088192 AAMC01088193 AAMC01088194 AAMC01088195 AAMC01088196 AAMC01088197 AAMC01088198 AAMC01088199 AAMC01088200 AKHW03000533 KYO45750.1 AHAT01001185 AHAT01001186 AHAT01001187 HAED01013205 HAEE01008207 SBQ99650.1 BC095763 AAH95763.1 HADZ01010707 HAEA01016296 SBP74648.1 HADY01006206 HAEJ01015231 SBP44691.1 HAEB01004392 SBQ50919.1 GBBI01003807 JAC14905.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000283053
UP000007151
+ More
UP000037510 UP000245037 UP000235965 UP000027135 UP000053105 UP000005205 UP000078540 UP000078492 UP000075809 UP000007755 UP000078542 UP000078541 UP000242457 UP000005203 UP000053825 UP000076502 UP000036403 UP000221080 UP000008237 UP000261440 UP000000311 UP000265080 UP000261580 UP000265100 UP000265160 UP000261460 UP000265000 UP000264840 UP000265040 UP000261400 UP000261660 UP000261360 UP000019118 UP000030742 UP000054308 UP000257160 UP000288216 UP000007635 UP000261420 UP000081671 UP000002852 UP000028760 UP000261480 UP000261500 UP000261640 UP000009046 UP000287033 UP000001038 UP000257200 UP000279307 UP000053097 UP000265180 UP000264800 UP000242638 UP000246464 UP000005207 UP000265020 UP000087266 UP000265200 UP000197619 UP000192220 UP000000539 UP000008143 UP000050525 UP000018468 UP000291020 UP000233080 UP000005226 UP000192223 UP000000437 UP000261540 UP000007303
UP000037510 UP000245037 UP000235965 UP000027135 UP000053105 UP000005205 UP000078540 UP000078492 UP000075809 UP000007755 UP000078542 UP000078541 UP000242457 UP000005203 UP000053825 UP000076502 UP000036403 UP000221080 UP000008237 UP000261440 UP000000311 UP000265080 UP000261580 UP000265100 UP000265160 UP000261460 UP000265000 UP000264840 UP000265040 UP000261400 UP000261660 UP000261360 UP000019118 UP000030742 UP000054308 UP000257160 UP000288216 UP000007635 UP000261420 UP000081671 UP000002852 UP000028760 UP000261480 UP000261500 UP000261640 UP000009046 UP000287033 UP000001038 UP000257200 UP000279307 UP000053097 UP000265180 UP000264800 UP000242638 UP000246464 UP000005207 UP000265020 UP000087266 UP000265200 UP000197619 UP000192220 UP000000539 UP000008143 UP000050525 UP000018468 UP000291020 UP000233080 UP000005226 UP000192223 UP000000437 UP000261540 UP000007303
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JCC8
S4NY43
A0A1E1VYH1
A0A0N1IER1
A0A194QG14
A0A2W1BK28
+ More
A0A2A4JX64 A0A3S2P5X2 A0A2H1VLK1 A0A212FCM4 A0A0L7KTD5 A0A2P8YJH0 A0A2J7R1B9 A0A067RBV0 A0A1B6CVB8 A0A1B6IXZ2 A0A1B6LMT0 A0A0M9A980 A0A158NM72 A0A195BWY4 A0A151JLI5 A0A151X1X5 F4WK14 A0A151IE89 A0A151JTN8 A0A2A3ENI4 A0A1B6GGX0 A0A088A105 A0A0L7QUJ3 E9J4J2 A0A154PSE5 A0A0J7KWA0 A0A2D0QAZ0 E2C2S7 A0A3B4C8D5 E3TD31 E2ABW1 A0A3P8S1C0 A0A3Q4HLZ3 A0A3P8PF05 A0A3P9DNQ3 A0A3B4FAU8 A0A3Q2Q9M6 A0A3Q2W676 A0A3Q1JUH6 A0A3B5B3E2 A0A3Q3GZQ0 A0A147A0T3 A0A3B4Y9S8 N6U9N4 A0A0F8ACM9 E6ZG47 A0A091I341 A0A3Q1C9B9 A0A401NYE2 G3P074 A0A3B4VMC3 A0A0S7HVT3 A0A1S3FTR9 A0A3B5RFN9 A0A087XLM6 A0A3B3XXJ9 A0A3B3U4K1 A0A3Q3MH21 E0VJI0 A0A1Y1L5E4 V9KWT3 A0A401SAA2 H2MQE0 A0A3Q1G422 A0A3L8DKV4 A0A026WGF9 A0A3P9KSA1 A0A3Q2ZAQ7 A0A3P9MVI9 A0A2U9BGI8 I3KTK6 A0A3Q2DPZ2 A0A3S2MCD1 A0A1S3L997 A0A3P9HFS6 A0A3B4VMC8 A0A218VCW3 A0A2I4BBN6 E1C1M8 F6R8M8 A0A151P9V2 W5MV18 A0A452IR59 A0A2K5ID15 A0A3B5JZ15 A0A1W4WHH3 A0A1A8IQP9 Q4VBI7 A0A1A8C6F6 A0A1A7ZQV6 A0A1A8EYI7 A0A023F083 A0A3B3RJ61 H3D095
A0A2A4JX64 A0A3S2P5X2 A0A2H1VLK1 A0A212FCM4 A0A0L7KTD5 A0A2P8YJH0 A0A2J7R1B9 A0A067RBV0 A0A1B6CVB8 A0A1B6IXZ2 A0A1B6LMT0 A0A0M9A980 A0A158NM72 A0A195BWY4 A0A151JLI5 A0A151X1X5 F4WK14 A0A151IE89 A0A151JTN8 A0A2A3ENI4 A0A1B6GGX0 A0A088A105 A0A0L7QUJ3 E9J4J2 A0A154PSE5 A0A0J7KWA0 A0A2D0QAZ0 E2C2S7 A0A3B4C8D5 E3TD31 E2ABW1 A0A3P8S1C0 A0A3Q4HLZ3 A0A3P8PF05 A0A3P9DNQ3 A0A3B4FAU8 A0A3Q2Q9M6 A0A3Q2W676 A0A3Q1JUH6 A0A3B5B3E2 A0A3Q3GZQ0 A0A147A0T3 A0A3B4Y9S8 N6U9N4 A0A0F8ACM9 E6ZG47 A0A091I341 A0A3Q1C9B9 A0A401NYE2 G3P074 A0A3B4VMC3 A0A0S7HVT3 A0A1S3FTR9 A0A3B5RFN9 A0A087XLM6 A0A3B3XXJ9 A0A3B3U4K1 A0A3Q3MH21 E0VJI0 A0A1Y1L5E4 V9KWT3 A0A401SAA2 H2MQE0 A0A3Q1G422 A0A3L8DKV4 A0A026WGF9 A0A3P9KSA1 A0A3Q2ZAQ7 A0A3P9MVI9 A0A2U9BGI8 I3KTK6 A0A3Q2DPZ2 A0A3S2MCD1 A0A1S3L997 A0A3P9HFS6 A0A3B4VMC8 A0A218VCW3 A0A2I4BBN6 E1C1M8 F6R8M8 A0A151P9V2 W5MV18 A0A452IR59 A0A2K5ID15 A0A3B5JZ15 A0A1W4WHH3 A0A1A8IQP9 Q4VBI7 A0A1A8C6F6 A0A1A7ZQV6 A0A1A8EYI7 A0A023F083 A0A3B3RJ61 H3D095
Ontologies
PATHWAY
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
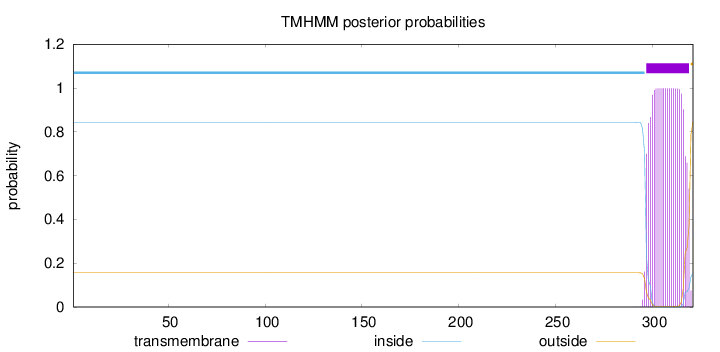
Length:
321
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.37588
Exp number, first 60 AAs:
0
Total prob of N-in:
0.84361
inside
1 - 296
TMhelix
297 - 319
outside
320 - 321
Population Genetic Test Statistics
Pi
15.686666
Theta
20.134026
Tajima's D
-0.624381
CLR
1.03819
CSRT
0.212189390530473
Interpretation
Uncertain
