Pre Gene Modal
BGIBMGA007173
Annotation
PREDICTED:_UPF0528_protein_CG10038_[Amyelois_transitella]
Full name
GPI inositol-deacylase
Location in the cell
Cytoplasmic Reliability : 1.292 Nuclear Reliability : 1.556
Sequence
CDS
ATGGAGGGAACGAAAACTATGAAGGAACTGGGATACATATTTGATTCAAATGGTCAAGTAAGAAAAATTGGAGCTGATGGACAGCCAACAGATGAACCATTCCAATTTGTTGTGAGCAATGATCACCAGGAATGCCAAGCTCATTATGAAGAACTTGGCTCAGCAATCACCAATTATGTTTATGATATTCTTCAGACTCAGATTGGATTGCAAAAGTTACCAATCCCAAAGGATTCAAATGATGGAACATTTATTTTTGTTTCAAAAGATTATGATAAAAAAGATGTTCTGTTAATATTGATTCACGGATCAGGCGTTGTGCGAGCTGGTCAGTGGGCTAGATCCCTCATAATCAATGAATCGTTGGACAAAGGATCTCAGATTCCGTATATCAAGAAGGCTTTGGCCAAAGACTATGGAGTAGTGGTACTAAATACAAATGACAATCACACAGCAGATGGAAAGACCATTCTCAACAGTAGCACCGCTGAGGAACATGCCCTTTACGCATGGCTGAACTACATATCCAATGCGAATGCAAGTTCCATACTCATTGTAGCACATAGTTATGGTGGGGTTGTCACGGTTTCTCTGGCAGAAGAGGTGCAAAATGACTTTGAAGACAGAGTAAAGGCCATTGCTTTTACTGACTCTGTACATTCCTATTCTAATAAGAAGCTTACAAACTTTTTGAAACAGGTTACAAAGAATTGGATATCTAGTCAGACACCGCTGGATACACCTCTCAGAACTCCTGATTTCGATATAACCAGACTTTCAGCAGGTCATGAGAAACATGAAATGACATCATACATGTGTATGGATTCTGTTTTTAAATACTTTGATGAAATACTAAATATGGATTAA
Protein
MEGTKTMKELGYIFDSNGQVRKIGADGQPTDEPFQFVVSNDHQECQAHYEELGSAITNYVYDILQTQIGLQKLPIPKDSNDGTFIFVSKDYDKKDVLLILIHGSGVVRAGQWARSLIINESLDKGSQIPYIKKALAKDYGVVVLNTNDNHTADGKTILNSSTAEEHALYAWLNYISNANASSILIVAHSYGGVVTVSLAEEVQNDFEDRVKAIAFTDSVHSYSNKKLTNFLKQVTKNWISSQTPLDTPLRTPDFDITRLSAGHEKHEMTSYMCMDSVFKYFDEILNMD
Summary
Description
Involved in inositol deacylation of GPI-anchored proteins which plays important roles in the quality control and ER-associated degradation of GPI-anchored proteins.
Similarity
Belongs to the GPI inositol-deacylase family.
Uniprot
H9JCC8
A0A2W1BJL6
A0A2A4JWV8
A0A2H1VLK1
A0A1E1W5T2
A0A3S2P5X2
+ More
S4PXI8 A0A0N1I822 A0A194QGE3 A0A0L7KTD5 A0A212FCM4 A0A224XTD7 A0A069DRP7 A0A023F8K8 R4FN09 A0A1B0GIA4 B0WSG8 A0A1Q3FCJ1 A0A1Q3FCP4 A0A411G7X9 A0A182TET4 A0A084VBK7 A0A182UYC7 A0A182Q5K1 A0A182IL43 A0A182P044 A0A182HSS0 A0A182YHQ2 Q7PGI6 F5HKW1 A0A182XAW6 A0A182LEV6 A0A131Y220 B7PYI4 A0A0L0CCF9 V5IES4 A0A182NRN4 A0A182K6J9 Q1HR15 W5J797 Q174E7 Q174E8 A0A1S4FEX9 A0A1S4FF73 A0A182FSP5 A0A336LLT8 A0A0P4VZ19 A0A0P4VU20 A0A2M4APV3 A0A2M4AQI9 A0A293LNW8 A0A2M4AQC6 A0A067RS02 A0A0P5SS43 A0A034W0P5 A0A0P5IST6 A0A162C7I6 T1P8P1 A0A182H049 A0A0N8ACB7 A0A0A1WK31 A0A0N8B1E9 A0A1I8MQ93 A0A034W3A3 A0A2B4RGB0 W8BVI7 A0A0P4XF55 A0A0A1WHB7 A0A1V9XJ35 A0A0N8EKF4 W8B7Y2 A0A0P5ES30 A0A1I8NS57 A0A2R5LD69 A0A1S4EHA6 W8C535 A0A0P6DXZ8 E9FWJ2 A0A0K8VZ49 A0A0P5M204 A0A182SA10 A0A131YVS0 A0A182RS50 A0A224YFQ4 A0A1A9VBG1 L7M4Q1 A0A1B0B3Q7 A0A1A9WJG5 R7UDD5 Q295Q8 A0A182VWV0 A0A0R3NIV0 B4GFU3 A0A1B0G5F0 A0A0L8I9T8 A0A182MVT1 A0A0P5YTH7 K1R8U3 A0A3B0KKX4 A0A0P5QIN2
S4PXI8 A0A0N1I822 A0A194QGE3 A0A0L7KTD5 A0A212FCM4 A0A224XTD7 A0A069DRP7 A0A023F8K8 R4FN09 A0A1B0GIA4 B0WSG8 A0A1Q3FCJ1 A0A1Q3FCP4 A0A411G7X9 A0A182TET4 A0A084VBK7 A0A182UYC7 A0A182Q5K1 A0A182IL43 A0A182P044 A0A182HSS0 A0A182YHQ2 Q7PGI6 F5HKW1 A0A182XAW6 A0A182LEV6 A0A131Y220 B7PYI4 A0A0L0CCF9 V5IES4 A0A182NRN4 A0A182K6J9 Q1HR15 W5J797 Q174E7 Q174E8 A0A1S4FEX9 A0A1S4FF73 A0A182FSP5 A0A336LLT8 A0A0P4VZ19 A0A0P4VU20 A0A2M4APV3 A0A2M4AQI9 A0A293LNW8 A0A2M4AQC6 A0A067RS02 A0A0P5SS43 A0A034W0P5 A0A0P5IST6 A0A162C7I6 T1P8P1 A0A182H049 A0A0N8ACB7 A0A0A1WK31 A0A0N8B1E9 A0A1I8MQ93 A0A034W3A3 A0A2B4RGB0 W8BVI7 A0A0P4XF55 A0A0A1WHB7 A0A1V9XJ35 A0A0N8EKF4 W8B7Y2 A0A0P5ES30 A0A1I8NS57 A0A2R5LD69 A0A1S4EHA6 W8C535 A0A0P6DXZ8 E9FWJ2 A0A0K8VZ49 A0A0P5M204 A0A182SA10 A0A131YVS0 A0A182RS50 A0A224YFQ4 A0A1A9VBG1 L7M4Q1 A0A1B0B3Q7 A0A1A9WJG5 R7UDD5 Q295Q8 A0A182VWV0 A0A0R3NIV0 B4GFU3 A0A1B0G5F0 A0A0L8I9T8 A0A182MVT1 A0A0P5YTH7 K1R8U3 A0A3B0KKX4 A0A0P5QIN2
EC Number
3.1.-.-
Pubmed
19121390
28756777
23622113
26354079
26227816
22118469
+ More
26334808 25474469 24438588 25244985 12364791 14747013 17210077 20966253 26108605 25765539 17204158 20920257 23761445 17510324 24845553 25348373 26483478 25830018 25315136 24495485 28327890 21292972 26830274 28797301 25576852 23254933 15632085 17994087 22992520
26334808 25474469 24438588 25244985 12364791 14747013 17210077 20966253 26108605 25765539 17204158 20920257 23761445 17510324 24845553 25348373 26483478 25830018 25315136 24495485 28327890 21292972 26830274 28797301 25576852 23254933 15632085 17994087 22992520
EMBL
BABH01031549
KZ150005
PZC75239.1
NWSH01000508
PCG75983.1
ODYU01002960
+ More
SOQ41124.1 GDQN01008740 JAT82314.1 RSAL01000317 RVE42595.1 GAIX01004963 JAA87597.1 KQ460882 KPJ11493.1 KQ458981 KPJ04489.1 JTDY01005870 KOB66532.1 AGBW02009157 OWR51496.1 GFTR01005053 JAW11373.1 GBGD01002141 JAC86748.1 GBBI01001022 JAC17690.1 ACPB03020702 GAHY01001514 JAA75996.1 AJWK01013619 DS232071 EDS33826.1 GFDL01009768 JAV25277.1 GFDL01009725 JAV25320.1 MH366192 QBB02001.1 ATLV01008177 KE524456 KFB35351.1 AXCN02000007 APCN01000336 AAAB01008859 EAA44919.5 EGK96864.1 EGK96863.1 GEFM01003274 JAP72522.1 ABJB010210014 DS820118 EEC11656.1 JRES01000608 KNC29940.1 GANP01009682 JAB74786.1 DQ440279 ABF18312.1 ADMH02002125 ETN58740.1 CH477412 EAT41447.1 EAT41446.1 UFQS01000012 UFQT01000012 SSW97227.1 SSX17613.1 GDRN01096866 GDRN01096864 JAI59255.1 GDRN01096869 GDRN01096868 GDRN01096867 GDRN01096863 GDRN01096862 JAI59257.1 GGFK01009505 MBW42826.1 GGFK01009720 MBW43041.1 GFWV01004788 MAA29518.1 GGFK01009663 MBW42984.1 KK852460 KDR23485.1 GDIP01136106 JAL67608.1 GAKP01010723 JAC48229.1 GDIQ01208466 JAK43259.1 LRGB01001361 KZS12442.1 KA645141 AFP59770.1 JXUM01100744 KQ564594 KXJ72041.1 GDIP01161401 JAJ62001.1 GBXI01014888 JAC99403.1 GDIQ01222267 JAK29458.1 GAKP01010724 JAC48228.1 LSMT01000529 PFX16651.1 GAMC01009229 JAB97326.1 GDIP01242010 JAI81391.1 GBXI01016035 JAC98256.1 MNPL01009920 OQR73451.1 GDIQ01018036 JAN76701.1 GAMC01009230 JAB97325.1 GDIP01144156 JAJ79246.1 GGLE01003249 MBY07375.1 GAMC01009231 JAB97324.1 GDIQ01070626 JAN24111.1 GL732526 EFX87903.1 GDHF01018842 GDHF01008469 JAI33472.1 JAI43845.1 GDIQ01164641 JAK87084.1 GEDV01005967 JAP82590.1 GFPF01004659 MAA15805.1 GACK01005718 JAA59316.1 JXJN01007968 AMQN01008283 KB302699 ELU03974.1 CM000070 EAL28650.3 KRT00914.1 CH479182 EDW34478.1 CCAG010000308 KQ416193 KOF98261.1 AXCM01005500 GDIP01067017 JAM36698.1 JH818603 EKC30411.1 OUUW01000008 SPP84438.1 GDIQ01134645 JAL17081.1
SOQ41124.1 GDQN01008740 JAT82314.1 RSAL01000317 RVE42595.1 GAIX01004963 JAA87597.1 KQ460882 KPJ11493.1 KQ458981 KPJ04489.1 JTDY01005870 KOB66532.1 AGBW02009157 OWR51496.1 GFTR01005053 JAW11373.1 GBGD01002141 JAC86748.1 GBBI01001022 JAC17690.1 ACPB03020702 GAHY01001514 JAA75996.1 AJWK01013619 DS232071 EDS33826.1 GFDL01009768 JAV25277.1 GFDL01009725 JAV25320.1 MH366192 QBB02001.1 ATLV01008177 KE524456 KFB35351.1 AXCN02000007 APCN01000336 AAAB01008859 EAA44919.5 EGK96864.1 EGK96863.1 GEFM01003274 JAP72522.1 ABJB010210014 DS820118 EEC11656.1 JRES01000608 KNC29940.1 GANP01009682 JAB74786.1 DQ440279 ABF18312.1 ADMH02002125 ETN58740.1 CH477412 EAT41447.1 EAT41446.1 UFQS01000012 UFQT01000012 SSW97227.1 SSX17613.1 GDRN01096866 GDRN01096864 JAI59255.1 GDRN01096869 GDRN01096868 GDRN01096867 GDRN01096863 GDRN01096862 JAI59257.1 GGFK01009505 MBW42826.1 GGFK01009720 MBW43041.1 GFWV01004788 MAA29518.1 GGFK01009663 MBW42984.1 KK852460 KDR23485.1 GDIP01136106 JAL67608.1 GAKP01010723 JAC48229.1 GDIQ01208466 JAK43259.1 LRGB01001361 KZS12442.1 KA645141 AFP59770.1 JXUM01100744 KQ564594 KXJ72041.1 GDIP01161401 JAJ62001.1 GBXI01014888 JAC99403.1 GDIQ01222267 JAK29458.1 GAKP01010724 JAC48228.1 LSMT01000529 PFX16651.1 GAMC01009229 JAB97326.1 GDIP01242010 JAI81391.1 GBXI01016035 JAC98256.1 MNPL01009920 OQR73451.1 GDIQ01018036 JAN76701.1 GAMC01009230 JAB97325.1 GDIP01144156 JAJ79246.1 GGLE01003249 MBY07375.1 GAMC01009231 JAB97324.1 GDIQ01070626 JAN24111.1 GL732526 EFX87903.1 GDHF01018842 GDHF01008469 JAI33472.1 JAI43845.1 GDIQ01164641 JAK87084.1 GEDV01005967 JAP82590.1 GFPF01004659 MAA15805.1 GACK01005718 JAA59316.1 JXJN01007968 AMQN01008283 KB302699 ELU03974.1 CM000070 EAL28650.3 KRT00914.1 CH479182 EDW34478.1 CCAG010000308 KQ416193 KOF98261.1 AXCM01005500 GDIP01067017 JAM36698.1 JH818603 EKC30411.1 OUUW01000008 SPP84438.1 GDIQ01134645 JAL17081.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000037510
+ More
UP000007151 UP000015103 UP000092461 UP000002320 UP000075902 UP000030765 UP000075903 UP000075886 UP000075880 UP000075885 UP000075840 UP000076408 UP000007062 UP000076407 UP000075882 UP000001555 UP000037069 UP000075884 UP000075881 UP000000673 UP000008820 UP000069272 UP000027135 UP000076858 UP000069940 UP000249989 UP000095301 UP000225706 UP000192247 UP000095300 UP000079169 UP000000305 UP000075901 UP000075900 UP000078200 UP000092460 UP000091820 UP000014760 UP000001819 UP000075920 UP000008744 UP000092444 UP000053454 UP000075883 UP000005408 UP000268350
UP000007151 UP000015103 UP000092461 UP000002320 UP000075902 UP000030765 UP000075903 UP000075886 UP000075880 UP000075885 UP000075840 UP000076408 UP000007062 UP000076407 UP000075882 UP000001555 UP000037069 UP000075884 UP000075881 UP000000673 UP000008820 UP000069272 UP000027135 UP000076858 UP000069940 UP000249989 UP000095301 UP000225706 UP000192247 UP000095300 UP000079169 UP000000305 UP000075901 UP000075900 UP000078200 UP000092460 UP000091820 UP000014760 UP000001819 UP000075920 UP000008744 UP000092444 UP000053454 UP000075883 UP000005408 UP000268350
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JCC8
A0A2W1BJL6
A0A2A4JWV8
A0A2H1VLK1
A0A1E1W5T2
A0A3S2P5X2
+ More
S4PXI8 A0A0N1I822 A0A194QGE3 A0A0L7KTD5 A0A212FCM4 A0A224XTD7 A0A069DRP7 A0A023F8K8 R4FN09 A0A1B0GIA4 B0WSG8 A0A1Q3FCJ1 A0A1Q3FCP4 A0A411G7X9 A0A182TET4 A0A084VBK7 A0A182UYC7 A0A182Q5K1 A0A182IL43 A0A182P044 A0A182HSS0 A0A182YHQ2 Q7PGI6 F5HKW1 A0A182XAW6 A0A182LEV6 A0A131Y220 B7PYI4 A0A0L0CCF9 V5IES4 A0A182NRN4 A0A182K6J9 Q1HR15 W5J797 Q174E7 Q174E8 A0A1S4FEX9 A0A1S4FF73 A0A182FSP5 A0A336LLT8 A0A0P4VZ19 A0A0P4VU20 A0A2M4APV3 A0A2M4AQI9 A0A293LNW8 A0A2M4AQC6 A0A067RS02 A0A0P5SS43 A0A034W0P5 A0A0P5IST6 A0A162C7I6 T1P8P1 A0A182H049 A0A0N8ACB7 A0A0A1WK31 A0A0N8B1E9 A0A1I8MQ93 A0A034W3A3 A0A2B4RGB0 W8BVI7 A0A0P4XF55 A0A0A1WHB7 A0A1V9XJ35 A0A0N8EKF4 W8B7Y2 A0A0P5ES30 A0A1I8NS57 A0A2R5LD69 A0A1S4EHA6 W8C535 A0A0P6DXZ8 E9FWJ2 A0A0K8VZ49 A0A0P5M204 A0A182SA10 A0A131YVS0 A0A182RS50 A0A224YFQ4 A0A1A9VBG1 L7M4Q1 A0A1B0B3Q7 A0A1A9WJG5 R7UDD5 Q295Q8 A0A182VWV0 A0A0R3NIV0 B4GFU3 A0A1B0G5F0 A0A0L8I9T8 A0A182MVT1 A0A0P5YTH7 K1R8U3 A0A3B0KKX4 A0A0P5QIN2
S4PXI8 A0A0N1I822 A0A194QGE3 A0A0L7KTD5 A0A212FCM4 A0A224XTD7 A0A069DRP7 A0A023F8K8 R4FN09 A0A1B0GIA4 B0WSG8 A0A1Q3FCJ1 A0A1Q3FCP4 A0A411G7X9 A0A182TET4 A0A084VBK7 A0A182UYC7 A0A182Q5K1 A0A182IL43 A0A182P044 A0A182HSS0 A0A182YHQ2 Q7PGI6 F5HKW1 A0A182XAW6 A0A182LEV6 A0A131Y220 B7PYI4 A0A0L0CCF9 V5IES4 A0A182NRN4 A0A182K6J9 Q1HR15 W5J797 Q174E7 Q174E8 A0A1S4FEX9 A0A1S4FF73 A0A182FSP5 A0A336LLT8 A0A0P4VZ19 A0A0P4VU20 A0A2M4APV3 A0A2M4AQI9 A0A293LNW8 A0A2M4AQC6 A0A067RS02 A0A0P5SS43 A0A034W0P5 A0A0P5IST6 A0A162C7I6 T1P8P1 A0A182H049 A0A0N8ACB7 A0A0A1WK31 A0A0N8B1E9 A0A1I8MQ93 A0A034W3A3 A0A2B4RGB0 W8BVI7 A0A0P4XF55 A0A0A1WHB7 A0A1V9XJ35 A0A0N8EKF4 W8B7Y2 A0A0P5ES30 A0A1I8NS57 A0A2R5LD69 A0A1S4EHA6 W8C535 A0A0P6DXZ8 E9FWJ2 A0A0K8VZ49 A0A0P5M204 A0A182SA10 A0A131YVS0 A0A182RS50 A0A224YFQ4 A0A1A9VBG1 L7M4Q1 A0A1B0B3Q7 A0A1A9WJG5 R7UDD5 Q295Q8 A0A182VWV0 A0A0R3NIV0 B4GFU3 A0A1B0G5F0 A0A0L8I9T8 A0A182MVT1 A0A0P5YTH7 K1R8U3 A0A3B0KKX4 A0A0P5QIN2
PDB
5IKK
E-value=2.05197e-05,
Score=113
Ontologies
GO
PANTHER
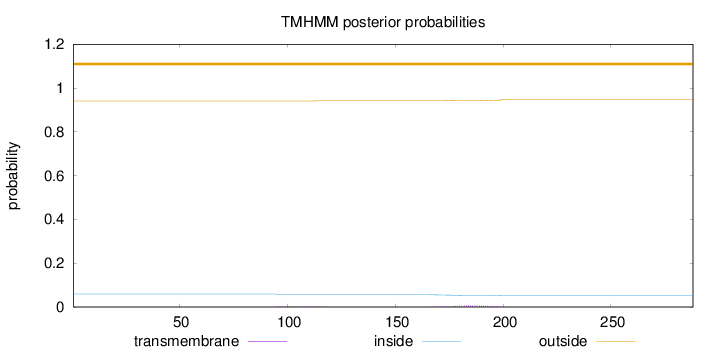
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
288
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21513
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.05960
outside
1 - 288
Population Genetic Test Statistics
Pi
14.008655
Theta
14.683272
Tajima's D
0.071377
CLR
0
CSRT
0.400729963501825
Interpretation
Uncertain
