Pre Gene Modal
BGIBMGA007175
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_YTHDC2_[Bombyx_mori]
Full name
3'-5' RNA helicase YTHDC2
Alternative Name
Keen to exit meiosis leaving testes under-populated protein
YTH domain-containing protein C2
YTH domain-containing protein 2
YTH domain-containing protein C2
YTH domain-containing protein 2
Location in the cell
Nuclear Reliability : 2.603
Sequence
CDS
ATGAGTTCTAAAAAGGGGAAACGAAGTTCTGGGAACAAACACTTCCCCATTGCGGAGTCAGTGAGCATAGCGCTGAAAATTCAGTTGGACAAGTTCATCAGTGATGACAACGAAACCGAACTTAAGTTTCCATCGTTCTTGTCGGCTCAAGAGCGCGGTTTTATTCATGAGACTGTTGTTAAATTAGGACTCAAATCGAAATCGCGCGGGAAAGGTGTCAACCGCTATTTAACAGTCTACAAAAGAGTAGGATCATCAATAATACAAAATGATGCAAAACTCATTTTGGACTCAAACATGAGGTGTAGCATTGCAGAGCTATCAAATACTTTTCCCATCACTAACAAAGAGAAGGATGATTTAAGCAGCTGTCCTGAAAAGGAAAGAAGCCCTCAACTCTCGCACAAGAGTTTGGGGCAGCTCAATAATGGAGTTGCACAGATTCCAAACACAACATACAATCCAGATTTATTGAAATTCAGAGAGAAACTTCCTGTTTATGAGCAGAGGCAAGATTTAATGAATGCCATCGTTCACAATCAGGTCATTATAGTAGCCGGAGCTACAGGGTGTGGAAAAACAACACAGCTTCCACAACTGGTGCTGGAGTATTGTCAACAACGTGGTCTTCCAGCCAGACTGTACTGCACCCAGCCGAGGAGAATTTCAGCTGTGTCTGTAGCCGAAAGGGTGGCGTATGAAAGGATGGAGAAAATAGGACAGTCAATTGGCTACCAGATCCGTCTGGAGTCGAGAGTCAGTCCTCGAACGGTGCTCACTTACTGTACCAACGGGGTGCTGCTCAGGACCCTCATGGGAGGAGACAGTGCACTTACTGGCGTGACCCACATACTGGTGGACGAGGTCCACGAGCGCGACAAGTTCAGCGACTTCCTGCTGATCGCCCTGAGGGACGCGCTGCCCAAGCACAAGGACCTCAAGCTCGTGCTCATGTCCGCCACCATGGACACGCAGATATTCTCCAGGTACTTCAACAACTGCCCGGTGATAACCATTCCGGGGCGGCTCCACGAGGTCCAGCGGTACTATCTCGAAGACGTACTGAAGATAACCAAATACAGAACGCCGAAAATGGTTCAAGTTGAAAAGGAACTGAAGAATAAATTGAAGTCTGGCAGGAGCTACTGGTCGCAGCTGGAGACTCAAAAAGCCGAAGACGGACCAAGCAAGGAAGTTAAAGAAGAAAAGGAGAATGTTCTGGATGCAGGTCTGCAGGCAGACATGGACGAGTTCATCGATGAATGCTTCAAGGAAGGCAGCTTGGACTCGTTCTGCCAGCTGCTGTACATGTACCTGTCGGAGGGCGTGCCCGTGCAGTGCGCGCACAGCCGCTCGGGCCGCTCGGGGCTCATGGCGGCGGCGGCGCGCGCCCTGCCCGAGACGCTGCACCAGCTGCTGCACATGGGTGCCGATCCATCCGCCAAAGACAAAGACGGCAAGACGGCATACGACTACGCCGTGGACAGCGGACACGCCGAGTCTGCCAAGCTACTCGCCACCTTCAACCTCACCGAGGACTCCAAGGAAAACCTGGAGGACAACGACGGCGACAACTTCCTCCTCGACGTCTACCACCACACCTTCTCCGAGGAACTCATAGACCACGATCTGATGCTGGCTCTCATAAAGTACATCCACGTCACGCTGCCGAAGGGGAGCATACTGGTGTTCCTCCCGGGCTACGACGACATAGTAACACTGAGGGACATGATCCAGTCTTGTCCGGAGATGAACGCCCAGAAGTACCAAATGTTCACGCTGCACAGCAACATGCAGACGGCCGACCAGAAGAAGGTGTTCAACGCGCTGCCCAACGCCAGGAAGATCATCGTCTCGACCAACATCGCGGAGACGTCCATCACGATCGACGACGTGGTGTTCGTGGTGGACTCGTGCAAGGTGAAGGAGAAGTACTACGAGTCCAACGGCGGCGTGTGCACGCTGCAGTGCGTGTGGAGCTCGCGGGCCTGCTGCCAGCAGCGCGCCGGCCGCGCGGGCAGGACCAAGCCCGGCCACTGCTTCCACATGTGCTCCAAGCGACGCTTCCAGACCCTGCCGCTCAACTCCGTGCCGGAGATCCTCCGCGTGCCCCTCCAGGAGCTGTGTCTGCACACCAAGCTGCTGGCGCCCGGGAACACGCCCATCGCTGACTTCCTGTCCAAAGCGTTGGAGCCTCCCTCGTTCCTGGCCGTCAGAAACGCGGTCACGTTACTGAAGACCATAGGGGCTCTGACCCCCATGGAGGACTTGACGGAGATCGGCCAACATCTACTAGACCTGACCGTGGAGCCCAAGTTAGGGAAGATGTTGCTTTACGCGTGCGTGATGAAGTGCCTCGACCCTATATTGACCATAGTGTGCAGTCTCGCGAATAAAGAACCTTTCCAAATATCTTTGAACCCTGAAAACAGGAAGAGAGGAAGTGCGGCTCGAAAAGAGTTCGCAGCCGACAGCTACTCCGACCATATGGCCTTGTTAAGAGCTTTCCAAGCTTGGCAGACAGCGCGAGCGAATGGAAACGAAAGAGCTTTCTGTACGAAGAACCTTATATGCGGAGCCACCATGGAAATGATCGTGGGATACAGATCTCAATTGCTGGCCCAATTAAGAGCACTGGGACTCGTCAAAGCTCGAGGATCGGGAGACATCAAAGACGTAAACTTGAACTCTGAGAAGTGGCATGTGGTGAAAGCTGTGCTAGTTAGCGGCCTGTATCCGTCCATAGCCCGCGTGGACAGGGACACCGGTACTTTGCGAACTTCGAAAGAAGTCAAAGTGGCATTCCATCCCAGTTCCACTTTGCACAAGGGCGGCGGTATCACGGGCTCCCAAAAATCGGTCATCAATGTTCCTACCGACTGGGTCGTGTTCGAAGAGATCTCGCGAGCGGGACGGTTCTGTTTCATTCGGTGCAACACCCTGGTGACGCCGCTGACCGTGGCGCTGTTCGGCGGCCCGCTGCGGCTGCCCCCCGGGGCGCTCACCCAGCGCGCCAACCCCCCGGGGCTGTCCAGCGACTCGGACAGCGAGGCCGACGAGAGCAACGCGTCCCCGGACACGGCCGTGCTCGCGCTGGACGACTGGATGGCCTTCACGGCAGACGCGTCCGACGCCATCAGCGTTTGCTACCTGCGACAGAAGCTCTGCGCTTTGGTCATCAGACGCATGTCGAACCCGGCCAAGCCCACGACGCCCCTGGACGACCAGATCCTGAACACCGTCGTCCATATCCTCGGAGCCGAAGAGAAGAACGTGGGCTTGAGTCAACCGAGCGGCATCGGCCAACGTCCCAAACCCTTGACCTTAGACTCGCCAAACTGGAGAATGAGGGTGGAAGAAGATCAGTACAGACACGACAAGTACTACCCGCAGTACAGCCAGCCGGAGTACCAGAATACCTTCTACCCGAACTACTTCAGGAACGGCCAGGGAAGGCCCGGCTGGAGGGGCGACGTGATGCCTCAAGGCTTCTCGCCGCAGTACAATGGCCCTAAGAGCCCTATGTCTCCGAACGGAAAGGTCATGATGCCCAACATGGAGAAGTACGGCGAGGGCGCGGAGGGGAACGCGAAGCAGTTCATGATAAAGGTGGACGAGCTGCACGCGCTGGAGGCCGCCTACAGCACGGGCAGCTACGTGTTCGCGCAGAACATCGCCAAAAGACTGCAGAAAGCTAAAATGGAGGGTCATCGTGTAACAGTATTCTTCTCGTGTCCGAGCGCCAACAAGTTCGTGGGCTGCGCGAACCTTCTGGACGTCAGCGGAGCCCTAGAGTGGATCTCCACGCAACACGTGCCTTATCATATGGTTCGACACTCGTGCGGGGGGGCGCGGGGCTGCGTGTGGCGCGAGGGGGAGCTGTGCGGCGCGGGGGGCGGCGCCCTGCTGGCGCACAGCCGGCGCCGGCACGCGCGACACCCGCACTGA
Protein
MSSKKGKRSSGNKHFPIAESVSIALKIQLDKFISDDNETELKFPSFLSAQERGFIHETVVKLGLKSKSRGKGVNRYLTVYKRVGSSIIQNDAKLILDSNMRCSIAELSNTFPITNKEKDDLSSCPEKERSPQLSHKSLGQLNNGVAQIPNTTYNPDLLKFREKLPVYEQRQDLMNAIVHNQVIIVAGATGCGKTTQLPQLVLEYCQQRGLPARLYCTQPRRISAVSVAERVAYERMEKIGQSIGYQIRLESRVSPRTVLTYCTNGVLLRTLMGGDSALTGVTHILVDEVHERDKFSDFLLIALRDALPKHKDLKLVLMSATMDTQIFSRYFNNCPVITIPGRLHEVQRYYLEDVLKITKYRTPKMVQVEKELKNKLKSGRSYWSQLETQKAEDGPSKEVKEEKENVLDAGLQADMDEFIDECFKEGSLDSFCQLLYMYLSEGVPVQCAHSRSGRSGLMAAAARALPETLHQLLHMGADPSAKDKDGKTAYDYAVDSGHAESAKLLATFNLTEDSKENLEDNDGDNFLLDVYHHTFSEELIDHDLMLALIKYIHVTLPKGSILVFLPGYDDIVTLRDMIQSCPEMNAQKYQMFTLHSNMQTADQKKVFNALPNARKIIVSTNIAETSITIDDVVFVVDSCKVKEKYYESNGGVCTLQCVWSSRACCQQRAGRAGRTKPGHCFHMCSKRRFQTLPLNSVPEILRVPLQELCLHTKLLAPGNTPIADFLSKALEPPSFLAVRNAVTLLKTIGALTPMEDLTEIGQHLLDLTVEPKLGKMLLYACVMKCLDPILTIVCSLANKEPFQISLNPENRKRGSAARKEFAADSYSDHMALLRAFQAWQTARANGNERAFCTKNLICGATMEMIVGYRSQLLAQLRALGLVKARGSGDIKDVNLNSEKWHVVKAVLVSGLYPSIARVDRDTGTLRTSKEVKVAFHPSSTLHKGGGITGSQKSVINVPTDWVVFEEISRAGRFCFIRCNTLVTPLTVALFGGPLRLPPGALTQRANPPGLSSDSDSEADESNASPDTAVLALDDWMAFTADASDAISVCYLRQKLCALVIRRMSNPAKPTTPLDDQILNTVVHILGAEEKNVGLSQPSGIGQRPKPLTLDSPNWRMRVEEDQYRHDKYYPQYSQPEYQNTFYPNYFRNGQGRPGWRGDVMPQGFSPQYNGPKSPMSPNGKVMMPNMEKYGEGAEGNAKQFMIKVDELHALEAAYSTGSYVFAQNIAKRLQKAKMEGHRVTVFFSCPSANKFVGCANLLDVSGALEWISTQHVPYHMVRHSCGGARGCVWREGELCGAGGGALLAHSRRRHARHPH
Summary
Description
3'-5' RNA helicase that plays a key role in the male and female germline by promoting transition from mitotic to meiotic divisions in stem cells (PubMed:28380054, PubMed:28809393, PubMed:29087293, PubMed:29033321, PubMed:29360036). Specifically recognizes and binds N6-methyladenosine (m6A)-containing RNAs, a modification present at internal sites of mRNAs and some non-coding RNAs that plays a role in the efficiency of RNA processing and stability (PubMed:29360036). Essential for ensuring a successful progression of the meiotic program in the germline by regulating the level of m6A-containing RNAs (PubMed:29033321). Acts by binding and promoting degradation of m6A-containing mRNAs: the 3'-5' RNA helicase activity is required for this process and RNA degradation may be mediated by XRN1 exoribonuclease (PubMed:29033321). Required for both spermatogenesis and oogenesis (PubMed:28809393, PubMed:29033321).
3'-5' RNA helicase that plays a key role in the male and female germline by promoting transition from mitotic to meiotic divisions in stem cells (PubMed:26318451, PubMed:29033321). Specifically recognizes and binds N6-methyladenosine (m6A)-containing RNAs, a modification present at internal sites of mRNAs and some non-coding RNAs that plays a role in the efficiency of RNA processing and stability (PubMed:26318451, PubMed:29033321). Essential for ensuring a successful progression of the meiotic program in the germline by regulating the level of m6A-containing RNAs (By similarity). Acts by binding and promoting degradation of m6A-containing mRNAs: the 3'-5' RNA helicase activity is required for this process and RNA degradation may be mediated by XRN1 exoribonuclease (PubMed:29033321). Required for both spermatogenesis and oogenesis (By similarity).
3'-5' RNA helicase that plays a key role in the male and female germline by promoting transition from mitotic to meiotic divisions in stem cells (PubMed:26318451, PubMed:29033321). Specifically recognizes and binds N6-methyladenosine (m6A)-containing RNAs, a modification present at internal sites of mRNAs and some non-coding RNAs that plays a role in the efficiency of RNA processing and stability (PubMed:26318451, PubMed:29033321). Essential for ensuring a successful progression of the meiotic program in the germline by regulating the level of m6A-containing RNAs (By similarity). Acts by binding and promoting degradation of m6A-containing mRNAs: the 3'-5' RNA helicase activity is required for this process and RNA degradation may be mediated by XRN1 exoribonuclease (PubMed:29033321). Required for both spermatogenesis and oogenesis (By similarity).
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Interacts with MEIOC; binds transcripts that regulate the mitotic cell cycle inhibiting progression into metaphase, thereby allowing meiotic prophase to proceed normally (PubMed:28380054, PubMed:29087293). Interacts (via ANK repeats) with XRN1 (By similarity).
Interacts with MEIOC; binds transcripts that regulate the mitotic cell cycle inhibiting progression into metaphase, thereby allowing meiotic prophase to proceed normally (By similarity). Interacts (via ANK repeats) with XRN1 (PubMed:29033321).
Interacts with MEIOC; binds transcripts that regulate the mitotic cell cycle inhibiting progression into metaphase, thereby allowing meiotic prophase to proceed normally (By similarity). Interacts (via ANK repeats) with XRN1 (PubMed:29033321).
Miscellaneous
'Ketu' is a harbinger of misfortune in Vedic mythology (PubMed:29360036).
Similarity
Belongs to the DEAD box helicase family. DEAH subfamily.
Keywords
ANK repeat
ATP-binding
Complete proteome
Cytoplasm
Differentiation
Helicase
Hydrolase
Meiosis
Nucleotide-binding
Oogenesis
Phosphoprotein
Reference proteome
Repeat
RNA-binding
Spermatogenesis
3D-structure
Polymorphism
Feature
chain 3'-5' RNA helicase YTHDC2
sequence variant In dbSNP:rs10071816.
sequence variant In dbSNP:rs10071816.
Uniprot
A0A2A4JX34
A0A2H1VJZ5
H9JCD0
A0A2W1BLT1
A0A0N0PBM8
A0A3S2LRW8
+ More
A0A194QM09 E2A5W6 A0A2J7RD30 A0A2J7RD43 A0A2J7RD34 E2BZ93 A0A067QLZ2 A0A026WKK0 A0A195B0M1 A0A3L8E2U3 A0A151WPU0 A0A158NY63 A0A151IJR0 A0A195EDS4 A0A195EUP5 K7J6R1 A0A088AKS3 A0A2A3EPV6 A0A2J7RD35 A0A0C9PKA5 T1JES8 A0A310SGF5 A0A0J7L5H3 E0VS89 A0A401SLK3 A0A401NIW9 A0A402FDR7 A0A2Y9DLV9 G1P6A1 A0A2Y9R2L8 H0UTJ8 A0A2Y9KSD4 A0A2Y9R7I4 L8I4A2 F1MNU7 A0A2Y9SD11 L5KLX8 A0A151PI96 A0A3Q7TND0 F6WCS8 W5KNU3 F7EI44 A0A2Y9KUT4 A0A452G8P8 A0A287AYE2 G3VK26 B2RR83 H3AS52 A0A2Y9I190 H9GD25 A0A3Q2H552 A0A3Q7WPM0 A0A3Q2HYT3 A0A1B6M6V0 A0A2U3XUL3 M3WVZ7 H0WJ02 G1LJ67 A0A2I2U0I4 A0A210QU91 A0A1V4JKF4 A0A3P9A718 A0A341BAA8 A0A384AYM1 A0A341BBS6 A0A2K6T3F9 A0A2U4BTH0 A0A2Y9PTT7 A0A2U4BT53 A0A2Y9PTV0 A0A3Q7PKV0 A0A1S3KZL1 A0A2Y9QWI3 A0A340WUJ7 S7PIX4 A0A0D9RQW7 A0A2U3WXR1 L5ME34 G3RES7 F7BPX9 K7G944 A0A3Q0GST4 A0A452HE79 Q9H6S0 A0A2K6DHY3 A0A3Q1N0J8 A0A2K5SGU5 H9YWL7 A0A452HE84 G1RLV7 A0A2J8XEY6 M3XNV2
A0A194QM09 E2A5W6 A0A2J7RD30 A0A2J7RD43 A0A2J7RD34 E2BZ93 A0A067QLZ2 A0A026WKK0 A0A195B0M1 A0A3L8E2U3 A0A151WPU0 A0A158NY63 A0A151IJR0 A0A195EDS4 A0A195EUP5 K7J6R1 A0A088AKS3 A0A2A3EPV6 A0A2J7RD35 A0A0C9PKA5 T1JES8 A0A310SGF5 A0A0J7L5H3 E0VS89 A0A401SLK3 A0A401NIW9 A0A402FDR7 A0A2Y9DLV9 G1P6A1 A0A2Y9R2L8 H0UTJ8 A0A2Y9KSD4 A0A2Y9R7I4 L8I4A2 F1MNU7 A0A2Y9SD11 L5KLX8 A0A151PI96 A0A3Q7TND0 F6WCS8 W5KNU3 F7EI44 A0A2Y9KUT4 A0A452G8P8 A0A287AYE2 G3VK26 B2RR83 H3AS52 A0A2Y9I190 H9GD25 A0A3Q2H552 A0A3Q7WPM0 A0A3Q2HYT3 A0A1B6M6V0 A0A2U3XUL3 M3WVZ7 H0WJ02 G1LJ67 A0A2I2U0I4 A0A210QU91 A0A1V4JKF4 A0A3P9A718 A0A341BAA8 A0A384AYM1 A0A341BBS6 A0A2K6T3F9 A0A2U4BTH0 A0A2Y9PTT7 A0A2U4BT53 A0A2Y9PTV0 A0A3Q7PKV0 A0A1S3KZL1 A0A2Y9QWI3 A0A340WUJ7 S7PIX4 A0A0D9RQW7 A0A2U3WXR1 L5ME34 G3RES7 F7BPX9 K7G944 A0A3Q0GST4 A0A452HE79 Q9H6S0 A0A2K6DHY3 A0A3Q1N0J8 A0A2K5SGU5 H9YWL7 A0A452HE84 G1RLV7 A0A2J8XEY6 M3XNV2
EC Number
3.6.4.13
Pubmed
19121390
28756777
26354079
20798317
24845553
24508170
+ More
30249741 21347285 20075255 20566863 30297745 21993624 22751099 19393038 23258410 22293439 19892987 25329095 17495919 21709235 15489334 17242355 21183079 28809393 28380054 29087293 29033321 29360036 9215903 21881562 17975172 20010809 28812685 25069045 22398555 25243066 17381049 28562605 15372022 14702039 18669648 19413330 19690332 21269460 21406692 23186163 26318451 25319552
30249741 21347285 20075255 20566863 30297745 21993624 22751099 19393038 23258410 22293439 19892987 25329095 17495919 21709235 15489334 17242355 21183079 28809393 28380054 29087293 29033321 29360036 9215903 21881562 17975172 20010809 28812685 25069045 22398555 25243066 17381049 28562605 15372022 14702039 18669648 19413330 19690332 21269460 21406692 23186163 26318451 25319552
EMBL
NWSH01000508
PCG75970.1
ODYU01002960
SOQ41126.1
BABH01031548
KZ150005
+ More
PZC75241.1 KQ460882 KPJ11495.1 RSAL01000317 RVE42597.1 KQ458981 KPJ04491.1 GL437072 EFN71147.1 NEVH01005303 PNF38741.1 PNF38747.1 PNF38745.1 GL451577 EFN78999.1 KK853168 KDR10374.1 KK107168 EZA56171.1 KQ976692 KYM77749.1 QOIP01000001 RLU26773.1 KQ982851 KYQ49884.1 ADTU01003242 KQ977355 KYN03316.1 KQ979074 KYN22994.1 KQ981958 KYN31980.1 AAZX01003118 KZ288195 PBC33795.1 PNF38746.1 GBYB01001463 GBYB01001464 JAG71230.1 JAG71231.1 JH432130 KQ764919 OAD54223.1 LBMM01000691 KMQ97744.1 DS235746 EEB16245.1 BEZZ01000354 GCC31285.1 BFAA01001144 GCB60814.1 BDOT01000203 GCF54497.1 AAPE02000278 AAPE02000279 AAKN02023000 AAKN02023001 AAKN02023002 AAKN02023003 AAKN02023004 AAKN02023005 AAKN02023006 JH882018 ELR51325.1 KB030673 ELK11568.1 AKHW03000179 KYO48816.1 LWLT01000010 AEMK02000012 AEFK01051448 AEFK01051449 AEFK01051450 AEFK01051451 BC138263 BC171951 AFYH01113949 AFYH01113950 AFYH01113951 AFYH01113952 AFYH01113953 AFYH01113954 AFYH01113955 AFYH01113956 AAWZ02001637 GEBQ01030145 GEBQ01008327 JAT09832.1 JAT31650.1 AANG04002524 AAQR03059537 AAQR03059538 AAQR03059539 AAQR03059540 AAQR03059541 AAQR03059542 AAQR03059543 AAQR03059544 ACTA01002088 ACTA01010088 ACTA01018088 ACTA01026088 ACTA01034088 NEDP02001843 OWF52301.1 LSYS01006904 OPJ72640.1 KE162363 EPQ07932.1 AQIB01043849 AQIB01043850 AQIB01043851 KB101591 ELK36515.1 CABD030040362 CABD030040363 CABD030040364 CABD030040365 GAMT01007047 GAMS01003066 GAMR01006947 GAMP01009530 JAB04814.1 JAB20070.1 JAB26985.1 JAB43225.1 AGCU01161868 AGCU01161869 AGCU01161870 AGCU01161871 AC093208 AC010389 BC137285 AK025593 JU471279 AFH28083.1 ADFV01187669 ADFV01187670 ADFV01187671 ADFV01187672 ADFV01187673 NDHI03003367 PNJ80468.1 AEYP01047868 AEYP01047869 AEYP01047870 AEYP01047871 AEYP01047872 AEYP01047873
PZC75241.1 KQ460882 KPJ11495.1 RSAL01000317 RVE42597.1 KQ458981 KPJ04491.1 GL437072 EFN71147.1 NEVH01005303 PNF38741.1 PNF38747.1 PNF38745.1 GL451577 EFN78999.1 KK853168 KDR10374.1 KK107168 EZA56171.1 KQ976692 KYM77749.1 QOIP01000001 RLU26773.1 KQ982851 KYQ49884.1 ADTU01003242 KQ977355 KYN03316.1 KQ979074 KYN22994.1 KQ981958 KYN31980.1 AAZX01003118 KZ288195 PBC33795.1 PNF38746.1 GBYB01001463 GBYB01001464 JAG71230.1 JAG71231.1 JH432130 KQ764919 OAD54223.1 LBMM01000691 KMQ97744.1 DS235746 EEB16245.1 BEZZ01000354 GCC31285.1 BFAA01001144 GCB60814.1 BDOT01000203 GCF54497.1 AAPE02000278 AAPE02000279 AAKN02023000 AAKN02023001 AAKN02023002 AAKN02023003 AAKN02023004 AAKN02023005 AAKN02023006 JH882018 ELR51325.1 KB030673 ELK11568.1 AKHW03000179 KYO48816.1 LWLT01000010 AEMK02000012 AEFK01051448 AEFK01051449 AEFK01051450 AEFK01051451 BC138263 BC171951 AFYH01113949 AFYH01113950 AFYH01113951 AFYH01113952 AFYH01113953 AFYH01113954 AFYH01113955 AFYH01113956 AAWZ02001637 GEBQ01030145 GEBQ01008327 JAT09832.1 JAT31650.1 AANG04002524 AAQR03059537 AAQR03059538 AAQR03059539 AAQR03059540 AAQR03059541 AAQR03059542 AAQR03059543 AAQR03059544 ACTA01002088 ACTA01010088 ACTA01018088 ACTA01026088 ACTA01034088 NEDP02001843 OWF52301.1 LSYS01006904 OPJ72640.1 KE162363 EPQ07932.1 AQIB01043849 AQIB01043850 AQIB01043851 KB101591 ELK36515.1 CABD030040362 CABD030040363 CABD030040364 CABD030040365 GAMT01007047 GAMS01003066 GAMR01006947 GAMP01009530 JAB04814.1 JAB20070.1 JAB26985.1 JAB43225.1 AGCU01161868 AGCU01161869 AGCU01161870 AGCU01161871 AC093208 AC010389 BC137285 AK025593 JU471279 AFH28083.1 ADFV01187669 ADFV01187670 ADFV01187671 ADFV01187672 ADFV01187673 NDHI03003367 PNJ80468.1 AEYP01047868 AEYP01047869 AEYP01047870 AEYP01047871 AEYP01047872 AEYP01047873
Proteomes
UP000218220
UP000005204
UP000053240
UP000283053
UP000053268
UP000000311
+ More
UP000235965 UP000008237 UP000027135 UP000053097 UP000078540 UP000279307 UP000075809 UP000005205 UP000078542 UP000078492 UP000078541 UP000002358 UP000005203 UP000242457 UP000036403 UP000009046 UP000287033 UP000288216 UP000288954 UP000248480 UP000001074 UP000005447 UP000248482 UP000009136 UP000248484 UP000010552 UP000050525 UP000286640 UP000002281 UP000018467 UP000002280 UP000291000 UP000008227 UP000007648 UP000000589 UP000008672 UP000248481 UP000001646 UP000286642 UP000245341 UP000011712 UP000005225 UP000008912 UP000242188 UP000190648 UP000265140 UP000252040 UP000261681 UP000233220 UP000245320 UP000248483 UP000286641 UP000087266 UP000265300 UP000029965 UP000245340 UP000001519 UP000007267 UP000189705 UP000291020 UP000005640 UP000233120 UP000233040 UP000001073 UP000000715
UP000235965 UP000008237 UP000027135 UP000053097 UP000078540 UP000279307 UP000075809 UP000005205 UP000078542 UP000078492 UP000078541 UP000002358 UP000005203 UP000242457 UP000036403 UP000009046 UP000287033 UP000288216 UP000288954 UP000248480 UP000001074 UP000005447 UP000248482 UP000009136 UP000248484 UP000010552 UP000050525 UP000286640 UP000002281 UP000018467 UP000002280 UP000291000 UP000008227 UP000007648 UP000000589 UP000008672 UP000248481 UP000001646 UP000286642 UP000245341 UP000011712 UP000005225 UP000008912 UP000242188 UP000190648 UP000265140 UP000252040 UP000261681 UP000233220 UP000245320 UP000248483 UP000286641 UP000087266 UP000265300 UP000029965 UP000245340 UP000001519 UP000007267 UP000189705 UP000291020 UP000005640 UP000233120 UP000233040 UP000001073 UP000000715
Pfam
Interpro
IPR007502
Helicase-assoc_dom
+ More
IPR031083 YTHDC2-related
IPR027417 P-loop_NTPase
IPR002110 Ankyrin_rpt
IPR036867 R3H_dom_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR011709 DUF1605
IPR020683 Ankyrin_rpt-contain_dom
IPR007275 YTH_domain
IPR011545 DEAD/DEAH_box_helicase_dom
IPR001374 R3H_dom
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR003593 AAA+_ATPase
IPR011704 ATPase_dyneun-rel_AAA
IPR002035 VWF_A
IPR039891 VWA8
IPR036465 vWFA_dom_sf
IPR029058 AB_hydrolase
IPR034083 R3H_DEXH_helicase
IPR031083 YTHDC2-related
IPR027417 P-loop_NTPase
IPR002110 Ankyrin_rpt
IPR036867 R3H_dom_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR011709 DUF1605
IPR020683 Ankyrin_rpt-contain_dom
IPR007275 YTH_domain
IPR011545 DEAD/DEAH_box_helicase_dom
IPR001374 R3H_dom
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR003593 AAA+_ATPase
IPR011704 ATPase_dyneun-rel_AAA
IPR002035 VWF_A
IPR039891 VWA8
IPR036465 vWFA_dom_sf
IPR029058 AB_hydrolase
IPR034083 R3H_DEXH_helicase
SUPFAM
ProteinModelPortal
A0A2A4JX34
A0A2H1VJZ5
H9JCD0
A0A2W1BLT1
A0A0N0PBM8
A0A3S2LRW8
+ More
A0A194QM09 E2A5W6 A0A2J7RD30 A0A2J7RD43 A0A2J7RD34 E2BZ93 A0A067QLZ2 A0A026WKK0 A0A195B0M1 A0A3L8E2U3 A0A151WPU0 A0A158NY63 A0A151IJR0 A0A195EDS4 A0A195EUP5 K7J6R1 A0A088AKS3 A0A2A3EPV6 A0A2J7RD35 A0A0C9PKA5 T1JES8 A0A310SGF5 A0A0J7L5H3 E0VS89 A0A401SLK3 A0A401NIW9 A0A402FDR7 A0A2Y9DLV9 G1P6A1 A0A2Y9R2L8 H0UTJ8 A0A2Y9KSD4 A0A2Y9R7I4 L8I4A2 F1MNU7 A0A2Y9SD11 L5KLX8 A0A151PI96 A0A3Q7TND0 F6WCS8 W5KNU3 F7EI44 A0A2Y9KUT4 A0A452G8P8 A0A287AYE2 G3VK26 B2RR83 H3AS52 A0A2Y9I190 H9GD25 A0A3Q2H552 A0A3Q7WPM0 A0A3Q2HYT3 A0A1B6M6V0 A0A2U3XUL3 M3WVZ7 H0WJ02 G1LJ67 A0A2I2U0I4 A0A210QU91 A0A1V4JKF4 A0A3P9A718 A0A341BAA8 A0A384AYM1 A0A341BBS6 A0A2K6T3F9 A0A2U4BTH0 A0A2Y9PTT7 A0A2U4BT53 A0A2Y9PTV0 A0A3Q7PKV0 A0A1S3KZL1 A0A2Y9QWI3 A0A340WUJ7 S7PIX4 A0A0D9RQW7 A0A2U3WXR1 L5ME34 G3RES7 F7BPX9 K7G944 A0A3Q0GST4 A0A452HE79 Q9H6S0 A0A2K6DHY3 A0A3Q1N0J8 A0A2K5SGU5 H9YWL7 A0A452HE84 G1RLV7 A0A2J8XEY6 M3XNV2
A0A194QM09 E2A5W6 A0A2J7RD30 A0A2J7RD43 A0A2J7RD34 E2BZ93 A0A067QLZ2 A0A026WKK0 A0A195B0M1 A0A3L8E2U3 A0A151WPU0 A0A158NY63 A0A151IJR0 A0A195EDS4 A0A195EUP5 K7J6R1 A0A088AKS3 A0A2A3EPV6 A0A2J7RD35 A0A0C9PKA5 T1JES8 A0A310SGF5 A0A0J7L5H3 E0VS89 A0A401SLK3 A0A401NIW9 A0A402FDR7 A0A2Y9DLV9 G1P6A1 A0A2Y9R2L8 H0UTJ8 A0A2Y9KSD4 A0A2Y9R7I4 L8I4A2 F1MNU7 A0A2Y9SD11 L5KLX8 A0A151PI96 A0A3Q7TND0 F6WCS8 W5KNU3 F7EI44 A0A2Y9KUT4 A0A452G8P8 A0A287AYE2 G3VK26 B2RR83 H3AS52 A0A2Y9I190 H9GD25 A0A3Q2H552 A0A3Q7WPM0 A0A3Q2HYT3 A0A1B6M6V0 A0A2U3XUL3 M3WVZ7 H0WJ02 G1LJ67 A0A2I2U0I4 A0A210QU91 A0A1V4JKF4 A0A3P9A718 A0A341BAA8 A0A384AYM1 A0A341BBS6 A0A2K6T3F9 A0A2U4BTH0 A0A2Y9PTT7 A0A2U4BT53 A0A2Y9PTV0 A0A3Q7PKV0 A0A1S3KZL1 A0A2Y9QWI3 A0A340WUJ7 S7PIX4 A0A0D9RQW7 A0A2U3WXR1 L5ME34 G3RES7 F7BPX9 K7G944 A0A3Q0GST4 A0A452HE79 Q9H6S0 A0A2K6DHY3 A0A3Q1N0J8 A0A2K5SGU5 H9YWL7 A0A452HE84 G1RLV7 A0A2J8XEY6 M3XNV2
PDB
5VHD
E-value=9.43334e-93,
Score=873
Ontologies
GO
PANTHER
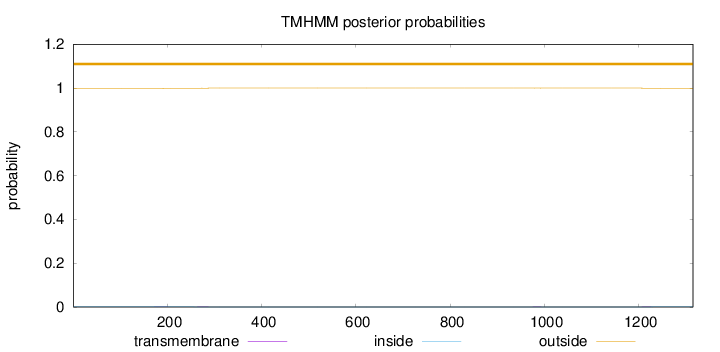
Topology
Subcellular location
Cytoplasm
Length:
1315
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0564000000000002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00163
outside
1 - 1315
Population Genetic Test Statistics
Pi
17.532909
Theta
14.486426
Tajima's D
-0.68379
CLR
1.590791
CSRT
0.206339683015849
Interpretation
Uncertain
