Gene
KWMTBOMO12730
Pre Gene Modal
BGIBMGA007191
Annotation
PREDICTED:_TFIIH_basal_transcription_factor_complex_helicase_XPD_subunit_isoform_X1_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 1.732
Sequence
CDS
ATGAAGCTAACTGTCGATGGTTTATTGGTCTATTTCCCATACGATTACATTTATCCGGAACAGTATGCTTATATGTTAGAGTTAAAACGTGCATTGGACGCTAAGGGACATGGATTGTTAGAGATGCCTTCCGGAACCGGAAAAACCATATCGCTCCTGTCTTTAATCGTAGCTTATATGATACAGAACCCTCATCATGTCAGAAAGTTAATCTACTGCTCTCGTACAGTGCCAGAAATAGAAAAAGTGTTAGAAGAACTAAAGAATTTATTCAATTATTATGAGAAATCTCAAGGAGAGAAACCGAACTTAACAGGAGTTGTTCTTAGTTCCAGGAAAAACTTATGTATTCATCCTGATGTATCAAGAGAAAGAGAAGGGAAGCTTGTAGATGGCAAATGTCATGCCCTCACAGCAAGCTATATACGAGACAGGCATGAACGTGATAGCTCTGTGCCAATTTGTCAATTTTATGAGGGCTTCAATCGTGAAGGCAAAGAGTCTATGTTACCATACGGAGTCTACACTATGGATGATATGAAACAATATGGTGCTGATAGAAATTGGTGTCCATATTTTCTTGCGAGATTTGCTATAATTCACGCTGAGATAGTGGTGTACTCATATCACTATTTGCTAGATCCAAAAATAGCAGAAGTTGTCTCGAAGGAACTGAACAGGGAGGCGGTGGTCGTGTTTGATGAGGCTCACAACATAGACAACGTGTGTATAGACTCCCTTAGTGTGAAGATCACGAGGCGGACCATAGACAAGAGCTCGCAGGCATTACAGCAACTTGAGAAGACTGTTGCAGAAATTCGTGACGAAGACGCGGCTCGTCTTACGATGGAGTACGAGCAACTGGTGGCAGGGCTGAAGGACGCCGCCATACAGAGGGAGACTGATACAATACTGGGCAACCCTATTTTACCCGATGAAGTGTTGAACGAGGTTGTACCCGGTAACATCAGGAACGCCGAGCACTTTCTCAGCTTCCTGAAGCGGTTCATCGAATATGTGAAGACCAGACTGCGCATACAGCATGTGGTGCAAGAGTCTCCTGCGGGTTTCTTAAAAGACGTATCGAGTCGGGTGTGCATAGAACGGAAGCCGCTACGATTCTGCGCATCTCGCCTCGCCTCCCTGATGAGAACGCTCCAGATACCGGATCCGTCGCACTTATCATCGCTAGTCCTGATCACGCATCTCGCGACCCTGGTCTCGACCTACACCAAGGGCTTCACGATCATAATCGAGCCCTTCGACGACAAAACGCCGAATGTTTCGAATCCTATATTGCATTTTTCATGTATGGACTCCTCAATAGCGATGCGGCCGGTCTTCGCGAGGTTCCAGACCGTCATCATCACGTCAGGAACCCTCTCGCCGCTGGACATGTACCCGAAGATCCTGGACTTCAATCCGGTGATAATGAGCTCCTTCACCATGACCCTGGCCAGGCCGTGCATACTGCCCATGATAGTATCGAAGGGCAGCGATCAGGTGGCCATATCGTCCAAGTACGAGACGAGAGAGGACGTCGCCGTCATAAGGAACTACGGGCAGCTGCTGGTCGAGATGTCGGCGTGCGTGCCTGACGGCGTGGTGTGCTTCTTCACGTCGTACCTGTATCTGGAGAGCGTCGTGGGGGCCTGGTACGACCAGGGTGTAGTCGCGAGTCTGCAGCGACACAAGCTGCTGTTCATCGAGACGCAAGACTCCGCCGAGACGAGCTTCGCCCTCATCAACTACATCAAGGCAAGTGGCGCCGCCGCCCCACCAGCGGGACCCTTACATATAGAGCATCACTAG
Protein
MKLTVDGLLVYFPYDYIYPEQYAYMLELKRALDAKGHGLLEMPSGTGKTISLLSLIVAYMIQNPHHVRKLIYCSRTVPEIEKVLEELKNLFNYYEKSQGEKPNLTGVVLSSRKNLCIHPDVSREREGKLVDGKCHALTASYIRDRHERDSSVPICQFYEGFNREGKESMLPYGVYTMDDMKQYGADRNWCPYFLARFAIIHAEIVVYSYHYLLDPKIAEVVSKELNREAVVVFDEAHNIDNVCIDSLSVKITRRTIDKSSQALQQLEKTVAEIRDEDAARLTMEYEQLVAGLKDAAIQRETDTILGNPILPDEVLNEVVPGNIRNAEHFLSFLKRFIEYVKTRLRIQHVVQESPAGFLKDVSSRVCIERKPLRFCASRLASLMRTLQIPDPSHLSSLVLITHLATLVSTYTKGFTIIIEPFDDKTPNVSNPILHFSCMDSSIAMRPVFARFQTVIITSGTLSPLDMYPKILDFNPVIMSSFTMTLARPCILPMIVSKGSDQVAISSKYETREDVAVIRNYGQLLVEMSACVPDGVVCFFTSYLYLESVVGAWYDQGVVASLQRHKLLFIETQDSAETSFALINYIKASGAAAPPAGPLHIEHH
Summary
Uniprot
A0A2A4JX44
A0A2W1BRD1
A0A2H1VL16
A0A194QG75
A0A212FCN4
A0A3S2NL39
+ More
A0A1Y1MDM5 A0A1Y1MHC2 A0A0L7QRM5 A0A088AL99 A0A2A3ES94 D6WI90 A0A195BVS4 A0A0M4ETK6 A0A1W4WWM3 N6UAL2 A0A151WNR8 F4WHJ1 B4KNC2 A0A0C9QGN2 A0A1W4UMC7 A0A151IBK0 A0A151K0U4 E2AAG2 B3NJP2 A0A0L0BUY2 B4P9G5 B4J6Q4 A0A3B0JLM5 B4MQM8 B5E1L9 B4H4V2 A0A151J393 A0A2J7PH21 A0A2J7PH38 A0A2J7PH36 A0A2J7PH26 Q7KVP9 D3DMG9 A0A067QTE7 B4LMQ6 B4QFN4 A0A2J7PH37 B4I7L3 B3MDT0 A0A2Z5U604 A0A1I8N808 A0A1B6EVK6 A0A0M8ZYN4 H9JCE6 A0A0A1X7Y6 A0A0K8UIG3 A0A034W8P2 A0A0K8VMZ9 A0A1I8Q5S7 A0A1B6E215 A0A026W2N3 A0A1B0CM32 A0A182V388 A0A1Q3G257 A0A182U4Q7 A0A182LFY1 A0A182WRM9 Q7QD77 A0A182HU49 A0A1A9Y0P4 A0A1B0BNJ1 A0A182N6B5 A0A182VRF5 A0A1A9ZWC9 Q0IGE4 A0A182IS41 A0A182QZS3 K7IS99 A0A182RT65 A0A182PE16 A0A1A9V8C9 A0A182F7Z9 W5J778 A0A154P6T6 A0A232EU60 A0A1A9WIP7 A0A182LWR4 A0A182XW15 A0A336K5U7 A0A2M4BEW6 A0A182T412 A0A1B0G7Q7 A0A182K4B3 A0A158N912 U4U951 A0A1J1J477 A0A310SJ47 A0A023F3L9 Q9XYZ2 A0A1B6LG37 E2BBM2 T1I4Q7 A0A0P6ABH5 A0A0P5YAY8
A0A1Y1MDM5 A0A1Y1MHC2 A0A0L7QRM5 A0A088AL99 A0A2A3ES94 D6WI90 A0A195BVS4 A0A0M4ETK6 A0A1W4WWM3 N6UAL2 A0A151WNR8 F4WHJ1 B4KNC2 A0A0C9QGN2 A0A1W4UMC7 A0A151IBK0 A0A151K0U4 E2AAG2 B3NJP2 A0A0L0BUY2 B4P9G5 B4J6Q4 A0A3B0JLM5 B4MQM8 B5E1L9 B4H4V2 A0A151J393 A0A2J7PH21 A0A2J7PH38 A0A2J7PH36 A0A2J7PH26 Q7KVP9 D3DMG9 A0A067QTE7 B4LMQ6 B4QFN4 A0A2J7PH37 B4I7L3 B3MDT0 A0A2Z5U604 A0A1I8N808 A0A1B6EVK6 A0A0M8ZYN4 H9JCE6 A0A0A1X7Y6 A0A0K8UIG3 A0A034W8P2 A0A0K8VMZ9 A0A1I8Q5S7 A0A1B6E215 A0A026W2N3 A0A1B0CM32 A0A182V388 A0A1Q3G257 A0A182U4Q7 A0A182LFY1 A0A182WRM9 Q7QD77 A0A182HU49 A0A1A9Y0P4 A0A1B0BNJ1 A0A182N6B5 A0A182VRF5 A0A1A9ZWC9 Q0IGE4 A0A182IS41 A0A182QZS3 K7IS99 A0A182RT65 A0A182PE16 A0A1A9V8C9 A0A182F7Z9 W5J778 A0A154P6T6 A0A232EU60 A0A1A9WIP7 A0A182LWR4 A0A182XW15 A0A336K5U7 A0A2M4BEW6 A0A182T412 A0A1B0G7Q7 A0A182K4B3 A0A158N912 U4U951 A0A1J1J477 A0A310SJ47 A0A023F3L9 Q9XYZ2 A0A1B6LG37 E2BBM2 T1I4Q7 A0A0P6ABH5 A0A0P5YAY8
Pubmed
28756777
26354079
22118469
28004739
18362917
19820115
+ More
23537049 21719571 17994087 20798317 26108605 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24845553 22936249 26760975 25315136 19121390 25830018 25348373 24508170 20966253 12364791 14747013 17210077 17510324 20075255 20920257 23761445 28648823 25244985 21347285 25474469
23537049 21719571 17994087 20798317 26108605 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24845553 22936249 26760975 25315136 19121390 25830018 25348373 24508170 20966253 12364791 14747013 17210077 17510324 20075255 20920257 23761445 28648823 25244985 21347285 25474469
EMBL
NWSH01000508
PCG75980.1
KZ150005
PZC75236.1
ODYU01002960
SOQ41122.1
+ More
KQ458981 KPJ04492.1 AGBW02009156 OWR51506.1 RSAL01000317 RVE42598.1 GEZM01036437 JAV82700.1 GEZM01036433 JAV82707.1 KQ414775 KOC61277.1 KZ288189 PBC34570.1 KQ971321 EFA00018.1 KQ976405 KYM91896.1 CP012524 ALC40862.1 APGK01042823 KB741007 KB632177 ENN75657.1 ERL89598.1 KQ982907 KYQ49458.1 GL888161 EGI66345.1 CH933808 EDW09975.2 GBYB01013738 GBYB01013739 JAG83505.1 JAG83506.1 KQ978095 KYM97017.1 KQ981229 KYN44334.1 GL438125 EFN69563.1 CH954179 EDV55421.1 JRES01001422 KNC23034.1 CM000158 EDW91285.1 CH916367 EDW00957.1 OUUW01000001 SPP74409.1 CH963849 EDW74417.1 CM000071 EDY69521.1 CH479210 EDW32788.1 KQ980290 KYN16909.1 NEVH01025150 PNF15635.1 PNF15639.1 PNF15640.1 PNF15637.1 AE013599 AAM70857.2 BT120038 ADA53577.1 KK852954 KDR13283.1 CH940648 EDW60997.1 CM000362 CM002911 EDX08026.1 KMY95458.1 PNF15641.1 CH480824 EDW56588.1 CH902619 EDV36465.2 FX985836 BBA93723.1 GECZ01027810 JAS41959.1 KQ435794 KOX73785.1 BABH01031546 BABH01031547 BABH01031548 GBXI01007050 JAD07242.1 GDHF01025986 JAI26328.1 GAKP01007878 JAC51074.1 GDHF01012101 JAI40213.1 GEDC01005357 JAS31941.1 KK107459 EZA50345.1 AJWK01018099 AJWK01018100 AJWK01018101 AJWK01018102 GFDL01001162 JAV33883.1 AAAB01008859 EAA08105.4 APCN01000606 JXJN01017417 CH477192 EAT48594.1 AXCN02000281 ADMH02002036 ETN59831.1 KQ434827 KZC07639.1 NNAY01002165 OXU21893.1 AXCM01003940 UFQS01000067 UFQT01000067 SSW98968.1 SSX19350.1 GGFJ01002431 MBW51572.1 CCAG010016890 ADTU01009023 KB632176 ERL89592.1 CVRI01000065 CRL05665.1 KQ762190 OAD56159.1 GBBI01002751 JAC15961.1 AF132140 AAD33587.1 GEBQ01017325 JAT22652.1 GL447128 EFN86899.1 ACPB03014607 GDIP01031692 JAM72023.1 GDIP01060392 LRGB01000868 JAM43323.1 KZS15446.1
KQ458981 KPJ04492.1 AGBW02009156 OWR51506.1 RSAL01000317 RVE42598.1 GEZM01036437 JAV82700.1 GEZM01036433 JAV82707.1 KQ414775 KOC61277.1 KZ288189 PBC34570.1 KQ971321 EFA00018.1 KQ976405 KYM91896.1 CP012524 ALC40862.1 APGK01042823 KB741007 KB632177 ENN75657.1 ERL89598.1 KQ982907 KYQ49458.1 GL888161 EGI66345.1 CH933808 EDW09975.2 GBYB01013738 GBYB01013739 JAG83505.1 JAG83506.1 KQ978095 KYM97017.1 KQ981229 KYN44334.1 GL438125 EFN69563.1 CH954179 EDV55421.1 JRES01001422 KNC23034.1 CM000158 EDW91285.1 CH916367 EDW00957.1 OUUW01000001 SPP74409.1 CH963849 EDW74417.1 CM000071 EDY69521.1 CH479210 EDW32788.1 KQ980290 KYN16909.1 NEVH01025150 PNF15635.1 PNF15639.1 PNF15640.1 PNF15637.1 AE013599 AAM70857.2 BT120038 ADA53577.1 KK852954 KDR13283.1 CH940648 EDW60997.1 CM000362 CM002911 EDX08026.1 KMY95458.1 PNF15641.1 CH480824 EDW56588.1 CH902619 EDV36465.2 FX985836 BBA93723.1 GECZ01027810 JAS41959.1 KQ435794 KOX73785.1 BABH01031546 BABH01031547 BABH01031548 GBXI01007050 JAD07242.1 GDHF01025986 JAI26328.1 GAKP01007878 JAC51074.1 GDHF01012101 JAI40213.1 GEDC01005357 JAS31941.1 KK107459 EZA50345.1 AJWK01018099 AJWK01018100 AJWK01018101 AJWK01018102 GFDL01001162 JAV33883.1 AAAB01008859 EAA08105.4 APCN01000606 JXJN01017417 CH477192 EAT48594.1 AXCN02000281 ADMH02002036 ETN59831.1 KQ434827 KZC07639.1 NNAY01002165 OXU21893.1 AXCM01003940 UFQS01000067 UFQT01000067 SSW98968.1 SSX19350.1 GGFJ01002431 MBW51572.1 CCAG010016890 ADTU01009023 KB632176 ERL89592.1 CVRI01000065 CRL05665.1 KQ762190 OAD56159.1 GBBI01002751 JAC15961.1 AF132140 AAD33587.1 GEBQ01017325 JAT22652.1 GL447128 EFN86899.1 ACPB03014607 GDIP01031692 JAM72023.1 GDIP01060392 LRGB01000868 JAM43323.1 KZS15446.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000283053
UP000053825
UP000005203
+ More
UP000242457 UP000007266 UP000078540 UP000092553 UP000192223 UP000019118 UP000030742 UP000075809 UP000007755 UP000009192 UP000192221 UP000078542 UP000078541 UP000000311 UP000008711 UP000037069 UP000002282 UP000001070 UP000268350 UP000007798 UP000001819 UP000008744 UP000078492 UP000235965 UP000000803 UP000027135 UP000008792 UP000000304 UP000001292 UP000007801 UP000095301 UP000053105 UP000005204 UP000095300 UP000053097 UP000092461 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000092443 UP000092460 UP000075884 UP000075920 UP000092445 UP000008820 UP000075880 UP000075886 UP000002358 UP000075900 UP000075885 UP000078200 UP000069272 UP000000673 UP000076502 UP000215335 UP000091820 UP000075883 UP000076408 UP000075901 UP000092444 UP000075881 UP000005205 UP000183832 UP000008237 UP000015103 UP000076858
UP000242457 UP000007266 UP000078540 UP000092553 UP000192223 UP000019118 UP000030742 UP000075809 UP000007755 UP000009192 UP000192221 UP000078542 UP000078541 UP000000311 UP000008711 UP000037069 UP000002282 UP000001070 UP000268350 UP000007798 UP000001819 UP000008744 UP000078492 UP000235965 UP000000803 UP000027135 UP000008792 UP000000304 UP000001292 UP000007801 UP000095301 UP000053105 UP000005204 UP000095300 UP000053097 UP000092461 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000092443 UP000092460 UP000075884 UP000075920 UP000092445 UP000008820 UP000075880 UP000075886 UP000002358 UP000075900 UP000075885 UP000078200 UP000069272 UP000000673 UP000076502 UP000215335 UP000091820 UP000075883 UP000076408 UP000075901 UP000092444 UP000075881 UP000005205 UP000183832 UP000008237 UP000015103 UP000076858
Interpro
IPR014013
Helic_SF1/SF2_ATP-bd_DinG/Rad3
+ More
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR006554 Helicase-like_DEXD_c2
IPR006555 ATP-dep_Helicase_C
IPR013020 Rad3/Chl1-like
IPR010643 HBB
IPR010614 DEAD_2
IPR001945 RAD3/XPD
IPR014001 Helicase_ATP-bd
IPR001202 WW_dom
IPR036020 WW_dom_sf
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR006554 Helicase-like_DEXD_c2
IPR006555 ATP-dep_Helicase_C
IPR013020 Rad3/Chl1-like
IPR010643 HBB
IPR010614 DEAD_2
IPR001945 RAD3/XPD
IPR014001 Helicase_ATP-bd
IPR001202 WW_dom
IPR036020 WW_dom_sf
CDD
ProteinModelPortal
A0A2A4JX44
A0A2W1BRD1
A0A2H1VL16
A0A194QG75
A0A212FCN4
A0A3S2NL39
+ More
A0A1Y1MDM5 A0A1Y1MHC2 A0A0L7QRM5 A0A088AL99 A0A2A3ES94 D6WI90 A0A195BVS4 A0A0M4ETK6 A0A1W4WWM3 N6UAL2 A0A151WNR8 F4WHJ1 B4KNC2 A0A0C9QGN2 A0A1W4UMC7 A0A151IBK0 A0A151K0U4 E2AAG2 B3NJP2 A0A0L0BUY2 B4P9G5 B4J6Q4 A0A3B0JLM5 B4MQM8 B5E1L9 B4H4V2 A0A151J393 A0A2J7PH21 A0A2J7PH38 A0A2J7PH36 A0A2J7PH26 Q7KVP9 D3DMG9 A0A067QTE7 B4LMQ6 B4QFN4 A0A2J7PH37 B4I7L3 B3MDT0 A0A2Z5U604 A0A1I8N808 A0A1B6EVK6 A0A0M8ZYN4 H9JCE6 A0A0A1X7Y6 A0A0K8UIG3 A0A034W8P2 A0A0K8VMZ9 A0A1I8Q5S7 A0A1B6E215 A0A026W2N3 A0A1B0CM32 A0A182V388 A0A1Q3G257 A0A182U4Q7 A0A182LFY1 A0A182WRM9 Q7QD77 A0A182HU49 A0A1A9Y0P4 A0A1B0BNJ1 A0A182N6B5 A0A182VRF5 A0A1A9ZWC9 Q0IGE4 A0A182IS41 A0A182QZS3 K7IS99 A0A182RT65 A0A182PE16 A0A1A9V8C9 A0A182F7Z9 W5J778 A0A154P6T6 A0A232EU60 A0A1A9WIP7 A0A182LWR4 A0A182XW15 A0A336K5U7 A0A2M4BEW6 A0A182T412 A0A1B0G7Q7 A0A182K4B3 A0A158N912 U4U951 A0A1J1J477 A0A310SJ47 A0A023F3L9 Q9XYZ2 A0A1B6LG37 E2BBM2 T1I4Q7 A0A0P6ABH5 A0A0P5YAY8
A0A1Y1MDM5 A0A1Y1MHC2 A0A0L7QRM5 A0A088AL99 A0A2A3ES94 D6WI90 A0A195BVS4 A0A0M4ETK6 A0A1W4WWM3 N6UAL2 A0A151WNR8 F4WHJ1 B4KNC2 A0A0C9QGN2 A0A1W4UMC7 A0A151IBK0 A0A151K0U4 E2AAG2 B3NJP2 A0A0L0BUY2 B4P9G5 B4J6Q4 A0A3B0JLM5 B4MQM8 B5E1L9 B4H4V2 A0A151J393 A0A2J7PH21 A0A2J7PH38 A0A2J7PH36 A0A2J7PH26 Q7KVP9 D3DMG9 A0A067QTE7 B4LMQ6 B4QFN4 A0A2J7PH37 B4I7L3 B3MDT0 A0A2Z5U604 A0A1I8N808 A0A1B6EVK6 A0A0M8ZYN4 H9JCE6 A0A0A1X7Y6 A0A0K8UIG3 A0A034W8P2 A0A0K8VMZ9 A0A1I8Q5S7 A0A1B6E215 A0A026W2N3 A0A1B0CM32 A0A182V388 A0A1Q3G257 A0A182U4Q7 A0A182LFY1 A0A182WRM9 Q7QD77 A0A182HU49 A0A1A9Y0P4 A0A1B0BNJ1 A0A182N6B5 A0A182VRF5 A0A1A9ZWC9 Q0IGE4 A0A182IS41 A0A182QZS3 K7IS99 A0A182RT65 A0A182PE16 A0A1A9V8C9 A0A182F7Z9 W5J778 A0A154P6T6 A0A232EU60 A0A1A9WIP7 A0A182LWR4 A0A182XW15 A0A336K5U7 A0A2M4BEW6 A0A182T412 A0A1B0G7Q7 A0A182K4B3 A0A158N912 U4U951 A0A1J1J477 A0A310SJ47 A0A023F3L9 Q9XYZ2 A0A1B6LG37 E2BBM2 T1I4Q7 A0A0P6ABH5 A0A0P5YAY8
PDB
6O9M
E-value=0,
Score=2203
Ontologies
GO
GO:0005524
GO:0006289
GO:0004003
GO:0003677
GO:0005634
GO:0004386
GO:0006468
GO:0006366
GO:0045893
GO:0000717
GO:0045951
GO:0006979
GO:0033683
GO:0000019
GO:0009411
GO:0003684
GO:0043141
GO:0000439
GO:0005703
GO:0005737
GO:0007348
GO:0005675
GO:0007088
GO:0051983
GO:0005705
GO:0010972
GO:0005700
GO:0006367
GO:0006974
GO:0001113
GO:0001111
GO:0016818
GO:0003676
GO:0008026
GO:0003707
GO:0016286
PANTHER
Topology
Subcellular location
Nucleus
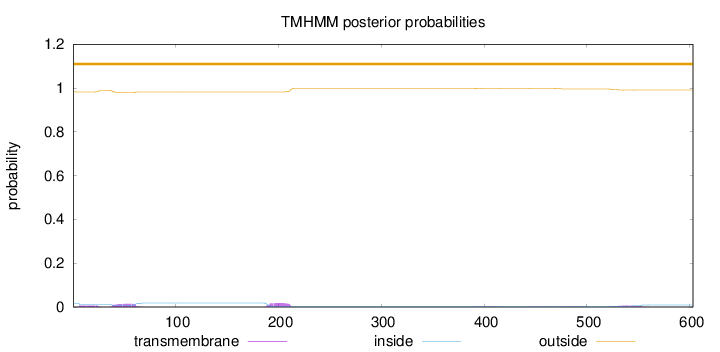
Length:
603
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.02424
Exp number, first 60 AAs:
0.40531
Total prob of N-in:
0.01748
outside
1 - 603
Population Genetic Test Statistics
Pi
18.290555
Theta
22.039107
Tajima's D
-0.941648
CLR
1.305272
CSRT
0.147642617869107
Interpretation
Uncertain
