Gene
KWMTBOMO12728
Pre Gene Modal
BGIBMGA007190
Annotation
PREDICTED:_D-amino-acid_oxidase_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.4
Sequence
CDS
ATGTTACAGATAGCTGTCGTGGGCGCCGGAATCAATGGCTTGACTTGCGCTTTGCGGATCCAAGAGAAATACAAGAAATATCGCGTCGTTCTATTAGCGAAACAATTCACCCCGAACACGACAGGGGACGGCTCCGCGGGTCTCTGGTACCCATTCGAAACGGGTAACACGTCCTCGGAATTATTATGTAAATGGGGTACTGCGACATACGAATTCCTGCACGGACTGTGGTTGGAGGGAGGCTTGGATGTATGTGCGGTGCCATTGTCCTTCGTCTATCGGAAGCCAAGAAACGAGAATAAGCCAGACTGGGGTAAACATACTTTCGGATATCGACAGATCGGAGAGAAGCACTTGGAGTATTTGAGCAAAAGGCATTCACAGAAATTCGTATCAGGCCATACCTTCACAACGCTAATTGTTCCACCTACAAAATTAATGGCACACTTTCACAAGCTGTTCGAAGAGGCTGGCGGAAGGACGCTTCAAGTGGAAGTGTCCTCGCTGGAGGACCCGATCTTGGGAGAATATGACGTGGTCGTGAACTGTACCGGGATTGGCGCGAGGGATCTGGTACCAGACAACAGCGTATTCTCTGTTAAAGGACAAGTGACTAAGGTATTCGCCCCTTGGGTCAACGAATGCATCGTGGACATCGACGGTGGAAATTACATCATACCTAATCCCGAAATATGTGTTCTGGGAGGGGTGACCGAGCACGGCAACTACAGCACCGACGTCGACGAGGACACGAGCGCGTTCATATTGAACGGTTGCAAAAAGATGTTGCCGGCGCTGCAGAACGTGGTCGTAGTGAACGAGTGGGCTGGTCTGAGGCCCGGCAGGGACCAGGTCAGGCTCGAAGCCGAGGAGAGGGGCGGGAAGTTGTACCTACACAACTATGGACACGGAGGCAGCGGCTTCACTCTGTTCTGGGGCTGCTCAACTGACGTCTTGTTCCTCTTGGAAAATTACTTAGAAAAGAAAGGAATAATAAAAAAATCGAAACTCTAA
Protein
MLQIAVVGAGINGLTCALRIQEKYKKYRVVLLAKQFTPNTTGDGSAGLWYPFETGNTSSELLCKWGTATYEFLHGLWLEGGLDVCAVPLSFVYRKPRNENKPDWGKHTFGYRQIGEKHLEYLSKRHSQKFVSGHTFTTLIVPPTKLMAHFHKLFEEAGGRTLQVEVSSLEDPILGEYDVVVNCTGIGARDLVPDNSVFSVKGQVTKVFAPWVNECIVDIDGGNYIIPNPEICVLGGVTEHGNYSTDVDEDTSAFILNGCKKMLPALQNVVVVNEWAGLRPGRDQVRLEAEERGGKLYLHNYGHGGSGFTLFWGCSTDVLFLLENYLEKKGIIKKSKL
Summary
Cofactor
FAD
Uniprot
H9JCE5
A0A194QG19
A0A2A4JW16
A0A0N1ICY8
A0A2W1BJI1
A0A3S2L063
+ More
I4DPR9 A0A212FCN0 A0A2H1VP54 B4JQK4 A0A034WPJ2 B4KJX8 A0A0K8V864 A0A182XNX4 A0A182L895 Q7PWY8 A0A182HK21 A0A182V1A4 A0A1D6TL57 A0A1J1ISD0 A0A2M4BT14 A0A2M3Z1R7 A0A1I8MJB8 A0A1W4UBY4 W8B4K5 A0A182JQN4 B4LRQ6 A0A182U9Y4 A0A1B0AK58 B4HXY3 A0A1A9UUS3 A0A1A9X595 B3MKQ4 A0A2M4CVV9 B4P006 B4N178 A0A182VW98 B3N5X6 A0A182YN98 A0A182R549 A0A182PR53 A0A1A9X8U6 A0A2M4ACP3 Q9VM80 B4Q525 A0A1B0AX58 A0A084WT56 A0A182H454 A0A2M4ACK8 Q29JZ1 B4GKY2 A0A1B0GFN9 A0A0M5J8W3 A0A182M103 A0A2M4ACM8 D3TL16 A0A3B0JL65 A0A1B0D4X5 A0A1S4EV77 Q17Q17 A0A336MZ36 M9NDG8 A0A1Q3FQM4 K7IXN6 A0A0L7QL03 A0A336M7F1 A0A0L0BR71 B0WJC4 A0A232FF57 A0A232EKL9 A0A2J7PTT6 A0A067RKT4 E2A1U5 D6WAM6 K7IR50 A0A1I8PRM8 A0A026X3J9 A0A1L8E2I9 A0A0M8ZS01 A0A1L8E2C6 A0A2M4CK89 A0A182SQS5 A0A1D2ND81 A0A195BEW0 A0A158NBX1 E9J689 A0A151J3C8 A0A195FK04 A0A226EVK3 T1GQ45 A0A0A1X8N8 A0A0K8TZN5 A0A182GZ97 A0A2A3E7E9 A0A088AUZ1 A0A0P6CTT2 E9GU05 A0A0P6D6M7 A0A0N8BCC3 A0A0N8AS57
I4DPR9 A0A212FCN0 A0A2H1VP54 B4JQK4 A0A034WPJ2 B4KJX8 A0A0K8V864 A0A182XNX4 A0A182L895 Q7PWY8 A0A182HK21 A0A182V1A4 A0A1D6TL57 A0A1J1ISD0 A0A2M4BT14 A0A2M3Z1R7 A0A1I8MJB8 A0A1W4UBY4 W8B4K5 A0A182JQN4 B4LRQ6 A0A182U9Y4 A0A1B0AK58 B4HXY3 A0A1A9UUS3 A0A1A9X595 B3MKQ4 A0A2M4CVV9 B4P006 B4N178 A0A182VW98 B3N5X6 A0A182YN98 A0A182R549 A0A182PR53 A0A1A9X8U6 A0A2M4ACP3 Q9VM80 B4Q525 A0A1B0AX58 A0A084WT56 A0A182H454 A0A2M4ACK8 Q29JZ1 B4GKY2 A0A1B0GFN9 A0A0M5J8W3 A0A182M103 A0A2M4ACM8 D3TL16 A0A3B0JL65 A0A1B0D4X5 A0A1S4EV77 Q17Q17 A0A336MZ36 M9NDG8 A0A1Q3FQM4 K7IXN6 A0A0L7QL03 A0A336M7F1 A0A0L0BR71 B0WJC4 A0A232FF57 A0A232EKL9 A0A2J7PTT6 A0A067RKT4 E2A1U5 D6WAM6 K7IR50 A0A1I8PRM8 A0A026X3J9 A0A1L8E2I9 A0A0M8ZS01 A0A1L8E2C6 A0A2M4CK89 A0A182SQS5 A0A1D2ND81 A0A195BEW0 A0A158NBX1 E9J689 A0A151J3C8 A0A195FK04 A0A226EVK3 T1GQ45 A0A0A1X8N8 A0A0K8TZN5 A0A182GZ97 A0A2A3E7E9 A0A088AUZ1 A0A0P6CTT2 E9GU05 A0A0P6D6M7 A0A0N8BCC3 A0A0N8AS57
Pubmed
19121390
26354079
28756777
22651552
22118469
17994087
+ More
25348373 20966253 12364791 14747013 17210077 25315136 24495485 17550304 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24438588 26483478 15632085 20353571 17510324 20075255 26108605 28648823 24845553 20798317 18362917 19820115 24508170 27289101 21347285 21282665 25830018 21292972
25348373 20966253 12364791 14747013 17210077 25315136 24495485 17550304 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24438588 26483478 15632085 20353571 17510324 20075255 26108605 28648823 24845553 20798317 18362917 19820115 24508170 27289101 21347285 21282665 25830018 21292972
EMBL
BABH01031544
BABH01031545
KQ458981
KPJ04493.1
NWSH01000508
PCG75976.1
+ More
KQ460882 KPJ11497.1 KZ150005 PZC75232.1 RSAL01000317 RVE42599.1 AK403745 BAM19909.1 AGBW02009156 OWR51505.1 ODYU01003620 SOQ42610.1 CH916372 EDV99184.1 GAKP01001436 JAC57516.1 CH933807 EDW12581.1 GDHF01017268 JAI35046.1 AAAB01008987 EAA01013.5 APCN01000814 CVRI01000058 CRL02618.1 GGFJ01006992 MBW56133.1 GGFM01001705 MBW22456.1 GAMC01014579 GAMC01014577 JAB91978.1 CH940649 EDW64658.1 CH480818 EDW51913.1 CH902620 EDV31585.1 GGFL01004790 MBW68968.1 CM000157 EDW87833.1 CH963920 EDW78018.1 CH954177 EDV59135.1 GGFK01005236 MBW38557.1 AE014134 AY089502 AAF52443.1 AAL90240.1 CM000361 CM002910 EDX04017.1 KMY88631.1 JXJN01005079 ATLV01026856 KE525420 KFB53400.1 JXUM01025363 JXUM01025364 JXUM01025365 KQ560720 KXJ81152.1 GGFK01005196 MBW38517.1 CH379062 EAL32820.1 CH479184 EDW37298.1 CCAG010011480 CP012523 ALC38494.1 AXCM01004139 GGFK01005151 MBW38472.1 EZ422109 ADD18311.1 OUUW01000006 SPP81573.1 AJVK01011634 CH477188 EAT48777.1 UFQT01004079 SSX35490.1 AFH03588.1 GFDL01005201 JAV29844.1 AAZX01012658 KQ414934 KOC59303.1 UFQT01000495 SSX24753.1 JRES01001480 KNC22575.1 DS231958 EDS29059.1 NNAY01000341 OXU29069.1 NNAY01003751 OXU18906.1 NEVH01021221 PNF19743.1 KK852619 KDR20119.1 GL435806 EFN72584.1 KQ971312 EEZ98651.1 KK107036 EZA61974.1 GFDF01001190 JAV12894.1 KQ435883 KOX69757.1 GFDF01001191 JAV12893.1 GGFL01001477 MBW65655.1 LJIJ01000088 ODN03045.1 KQ976500 KYM83113.1 ADTU01011441 GL768230 EFZ11661.1 KQ980286 KYN16939.1 KQ981491 KYN41000.1 LNIX01000001 OXA61612.1 CAQQ02161561 GBXI01007254 JAD07038.1 GDHF01032601 JAI19713.1 JXUM01099086 KQ564468 KXJ72229.1 KZ288356 PBC27212.1 GDIQ01119151 GDIQ01087415 JAN07322.1 GL732565 EFX77055.1 GDIQ01084113 JAN10624.1 GDIQ01191675 JAK60050.1 GDIQ01248203 GDIQ01084114 JAK03522.1
KQ460882 KPJ11497.1 KZ150005 PZC75232.1 RSAL01000317 RVE42599.1 AK403745 BAM19909.1 AGBW02009156 OWR51505.1 ODYU01003620 SOQ42610.1 CH916372 EDV99184.1 GAKP01001436 JAC57516.1 CH933807 EDW12581.1 GDHF01017268 JAI35046.1 AAAB01008987 EAA01013.5 APCN01000814 CVRI01000058 CRL02618.1 GGFJ01006992 MBW56133.1 GGFM01001705 MBW22456.1 GAMC01014579 GAMC01014577 JAB91978.1 CH940649 EDW64658.1 CH480818 EDW51913.1 CH902620 EDV31585.1 GGFL01004790 MBW68968.1 CM000157 EDW87833.1 CH963920 EDW78018.1 CH954177 EDV59135.1 GGFK01005236 MBW38557.1 AE014134 AY089502 AAF52443.1 AAL90240.1 CM000361 CM002910 EDX04017.1 KMY88631.1 JXJN01005079 ATLV01026856 KE525420 KFB53400.1 JXUM01025363 JXUM01025364 JXUM01025365 KQ560720 KXJ81152.1 GGFK01005196 MBW38517.1 CH379062 EAL32820.1 CH479184 EDW37298.1 CCAG010011480 CP012523 ALC38494.1 AXCM01004139 GGFK01005151 MBW38472.1 EZ422109 ADD18311.1 OUUW01000006 SPP81573.1 AJVK01011634 CH477188 EAT48777.1 UFQT01004079 SSX35490.1 AFH03588.1 GFDL01005201 JAV29844.1 AAZX01012658 KQ414934 KOC59303.1 UFQT01000495 SSX24753.1 JRES01001480 KNC22575.1 DS231958 EDS29059.1 NNAY01000341 OXU29069.1 NNAY01003751 OXU18906.1 NEVH01021221 PNF19743.1 KK852619 KDR20119.1 GL435806 EFN72584.1 KQ971312 EEZ98651.1 KK107036 EZA61974.1 GFDF01001190 JAV12894.1 KQ435883 KOX69757.1 GFDF01001191 JAV12893.1 GGFL01001477 MBW65655.1 LJIJ01000088 ODN03045.1 KQ976500 KYM83113.1 ADTU01011441 GL768230 EFZ11661.1 KQ980286 KYN16939.1 KQ981491 KYN41000.1 LNIX01000001 OXA61612.1 CAQQ02161561 GBXI01007254 JAD07038.1 GDHF01032601 JAI19713.1 JXUM01099086 KQ564468 KXJ72229.1 KZ288356 PBC27212.1 GDIQ01119151 GDIQ01087415 JAN07322.1 GL732565 EFX77055.1 GDIQ01084113 JAN10624.1 GDIQ01191675 JAK60050.1 GDIQ01248203 GDIQ01084114 JAK03522.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000283053
UP000007151
+ More
UP000001070 UP000009192 UP000076407 UP000075882 UP000007062 UP000075840 UP000075903 UP000183832 UP000095301 UP000192221 UP000075881 UP000008792 UP000075902 UP000092445 UP000001292 UP000078200 UP000091820 UP000007801 UP000002282 UP000007798 UP000075920 UP000008711 UP000076408 UP000075900 UP000075885 UP000092443 UP000000803 UP000000304 UP000092460 UP000030765 UP000069940 UP000249989 UP000001819 UP000008744 UP000092444 UP000092553 UP000075883 UP000268350 UP000092462 UP000008820 UP000002358 UP000053825 UP000037069 UP000002320 UP000215335 UP000235965 UP000027135 UP000000311 UP000007266 UP000095300 UP000053097 UP000053105 UP000075901 UP000094527 UP000078540 UP000005205 UP000078492 UP000078541 UP000198287 UP000015102 UP000242457 UP000005203 UP000000305
UP000001070 UP000009192 UP000076407 UP000075882 UP000007062 UP000075840 UP000075903 UP000183832 UP000095301 UP000192221 UP000075881 UP000008792 UP000075902 UP000092445 UP000001292 UP000078200 UP000091820 UP000007801 UP000002282 UP000007798 UP000075920 UP000008711 UP000076408 UP000075900 UP000075885 UP000092443 UP000000803 UP000000304 UP000092460 UP000030765 UP000069940 UP000249989 UP000001819 UP000008744 UP000092444 UP000092553 UP000075883 UP000268350 UP000092462 UP000008820 UP000002358 UP000053825 UP000037069 UP000002320 UP000215335 UP000235965 UP000027135 UP000000311 UP000007266 UP000095300 UP000053097 UP000053105 UP000075901 UP000094527 UP000078540 UP000005205 UP000078492 UP000078541 UP000198287 UP000015102 UP000242457 UP000005203 UP000000305
Pfam
PF01266 DAO
ProteinModelPortal
H9JCE5
A0A194QG19
A0A2A4JW16
A0A0N1ICY8
A0A2W1BJI1
A0A3S2L063
+ More
I4DPR9 A0A212FCN0 A0A2H1VP54 B4JQK4 A0A034WPJ2 B4KJX8 A0A0K8V864 A0A182XNX4 A0A182L895 Q7PWY8 A0A182HK21 A0A182V1A4 A0A1D6TL57 A0A1J1ISD0 A0A2M4BT14 A0A2M3Z1R7 A0A1I8MJB8 A0A1W4UBY4 W8B4K5 A0A182JQN4 B4LRQ6 A0A182U9Y4 A0A1B0AK58 B4HXY3 A0A1A9UUS3 A0A1A9X595 B3MKQ4 A0A2M4CVV9 B4P006 B4N178 A0A182VW98 B3N5X6 A0A182YN98 A0A182R549 A0A182PR53 A0A1A9X8U6 A0A2M4ACP3 Q9VM80 B4Q525 A0A1B0AX58 A0A084WT56 A0A182H454 A0A2M4ACK8 Q29JZ1 B4GKY2 A0A1B0GFN9 A0A0M5J8W3 A0A182M103 A0A2M4ACM8 D3TL16 A0A3B0JL65 A0A1B0D4X5 A0A1S4EV77 Q17Q17 A0A336MZ36 M9NDG8 A0A1Q3FQM4 K7IXN6 A0A0L7QL03 A0A336M7F1 A0A0L0BR71 B0WJC4 A0A232FF57 A0A232EKL9 A0A2J7PTT6 A0A067RKT4 E2A1U5 D6WAM6 K7IR50 A0A1I8PRM8 A0A026X3J9 A0A1L8E2I9 A0A0M8ZS01 A0A1L8E2C6 A0A2M4CK89 A0A182SQS5 A0A1D2ND81 A0A195BEW0 A0A158NBX1 E9J689 A0A151J3C8 A0A195FK04 A0A226EVK3 T1GQ45 A0A0A1X8N8 A0A0K8TZN5 A0A182GZ97 A0A2A3E7E9 A0A088AUZ1 A0A0P6CTT2 E9GU05 A0A0P6D6M7 A0A0N8BCC3 A0A0N8AS57
I4DPR9 A0A212FCN0 A0A2H1VP54 B4JQK4 A0A034WPJ2 B4KJX8 A0A0K8V864 A0A182XNX4 A0A182L895 Q7PWY8 A0A182HK21 A0A182V1A4 A0A1D6TL57 A0A1J1ISD0 A0A2M4BT14 A0A2M3Z1R7 A0A1I8MJB8 A0A1W4UBY4 W8B4K5 A0A182JQN4 B4LRQ6 A0A182U9Y4 A0A1B0AK58 B4HXY3 A0A1A9UUS3 A0A1A9X595 B3MKQ4 A0A2M4CVV9 B4P006 B4N178 A0A182VW98 B3N5X6 A0A182YN98 A0A182R549 A0A182PR53 A0A1A9X8U6 A0A2M4ACP3 Q9VM80 B4Q525 A0A1B0AX58 A0A084WT56 A0A182H454 A0A2M4ACK8 Q29JZ1 B4GKY2 A0A1B0GFN9 A0A0M5J8W3 A0A182M103 A0A2M4ACM8 D3TL16 A0A3B0JL65 A0A1B0D4X5 A0A1S4EV77 Q17Q17 A0A336MZ36 M9NDG8 A0A1Q3FQM4 K7IXN6 A0A0L7QL03 A0A336M7F1 A0A0L0BR71 B0WJC4 A0A232FF57 A0A232EKL9 A0A2J7PTT6 A0A067RKT4 E2A1U5 D6WAM6 K7IR50 A0A1I8PRM8 A0A026X3J9 A0A1L8E2I9 A0A0M8ZS01 A0A1L8E2C6 A0A2M4CK89 A0A182SQS5 A0A1D2ND81 A0A195BEW0 A0A158NBX1 E9J689 A0A151J3C8 A0A195FK04 A0A226EVK3 T1GQ45 A0A0A1X8N8 A0A0K8TZN5 A0A182GZ97 A0A2A3E7E9 A0A088AUZ1 A0A0P6CTT2 E9GU05 A0A0P6D6M7 A0A0N8BCC3 A0A0N8AS57
PDB
1VE9
E-value=1.7484e-39,
Score=408
Ontologies
PATHWAY
00260
Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00472 D-Arginine and D-ornithine metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04146 Peroxisome - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00472 D-Arginine and D-ornithine metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04146 Peroxisome - Bombyx mori (domestic silkworm)
GO
PANTHER
Topology
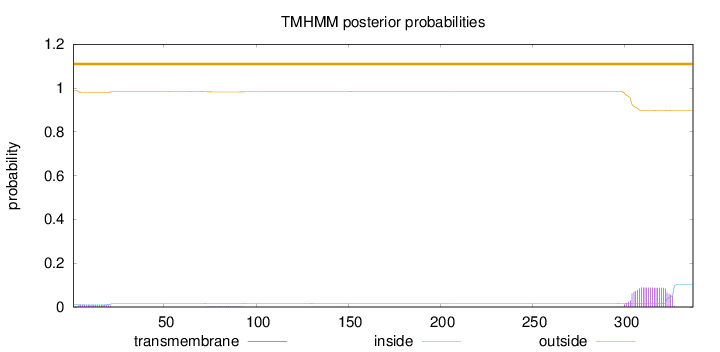
Length:
337
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.14733
Exp number, first 60 AAs:
0.17265
Total prob of N-in:
0.01228
outside
1 - 337
Population Genetic Test Statistics
Pi
5.797085
Theta
16.600658
Tajima's D
-1.382417
CLR
1.550199
CSRT
0.0739463026848658
Interpretation
Uncertain
