Gene
KWMTBOMO12724
Pre Gene Modal
BGIBMGA007181
Annotation
PREDICTED:_A-kinase_anchor_protein_17A_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.011
Sequence
CDS
ATGATCCAACCTGAGAGTTTTTCATTGTTAAAAGTGTCGAAACATAGTTCGGAAGTGATCAGATTTGATGCGGAAATAGAAAATTATAGTATGCTAAAGATGGTGATTATGAGACTTGATGAACGTGTGGTGAAGTTGAATGATTACCCGGAGCCCTTAAGAGTGAAAGTTTGTGAAGCGAAGTCGGATTTCCCTTCTCGTCACGCTTGGGACTCGTTCTTTCGAGATGCGTCCGATATGGATGAGATGAAGCCAGGAGAAAGACCAGACACTATTCATATATCAAACCTGCCAATACGTTGGTTTGTTCATAAAAATGAACGGGATGAAGATGCGATGCCTTCTGAGAGCTTATTCAAAAAGGTCTTTGAAAAGTTTGGTAGGGTACGTCAAGTGGACATACCTATTGCTGATCCATTTAGACTTCAAATGAAATCAGCCATCAGAGGCATCATGTATCCAACAGAAGATGCAGCTGTGTATTTTGAAGGATACGTACAGTTTAATGAATATGTCGGTTTCGTCAGATGTATGGATGAATTGAGAGGCAAGAAATTAGTTAGAAAGCAGGATGACATTGCCGAGTGGTGTAGCATCAGGGTTGATTTTGATAAGACAAAGCACATGACTGATGCAGCCATCAAGAGAAGGGCTATAGTTAGAGAAAGACTTGCAGCACGAAAGAGTCACAAAGAGGAAGAAGAACAAGCGGAAAAATTGAAAGCTGCTAAAAGAGAAGCAAAGGAAAGACAAAAATATGAATTGGCTGAAAAAGAGAAATTGGAGAAGATGAGGCAAAGAGAAGAAAGGAGAAAGAAAAAGCAACTTGCCAAACTAATGGAGAGAGACAATGTAGACCTGAATAGCAAAGTGGCAGAAGAACAGAAGAAGTTATTGAAAGCTCAAAAAAAGTTACAAGCCATTAGGCTAGTTGCAGAATTGTTTAAGAGAATTGAGCTCCGTCCAGAACTACAGCGTAATGGTGCTAAAACTCACAGAGCATACAATGAAAGCTTGAGGGCTCGTGTCGTAGAAAAGTTCAAAAGGCAACAAGAAGAATTGTTGGAAGCACAAAGGAAGAATCTCAGAAGCGCACTTGATGGTCGTATAGTGATCCGTTCAGCTTTAGAAACGAAGAAGCCGAGACGTACCTCTTCCCTCAGCTCTCTATCGGCTGACGAAGAAAATCAACTACGAATAAAAACAGAAAGAATCCCAAGTCCGGACTCGAGAATGCAATATGATAAAACACCTGTTCCTGCGATGTACGGGATGGCGAACCCTTACAGCTTCGGGTACCCGGCGTTCATGCCGCCAGGATATGCCTATGGACCGCAAAGTTCAATGTACTACCCCCGCGCGGCGTACCCTGCGGAGTGGGCGCGGCGCGCGGGCGGGGCGCGGGGGCGCGGGCGCGGCGTGCGGGGGCGCGGGGGGGCGCGGCCGCGGATGCCGTACTTCGACGGACCGGACGCCACCGCGCAGTACTACAGATACTTCAAGAAGCTGAAGGAATCCAACGAACACGACGACCGGCTGCATTCTCGCTCCCGCTCTCGGTCCCGCTCCCGCTCCCGCCGCCGCTCGTACACCCGCTCCCGCTCCCGCCGCCGCTCATACTCCAGGTCGAGGTCTCGTTCAAGGAGATCCAGATCTAGGAGTCGTTCCAGGAGTCGTTCAAAGAGCAGAAGCAGACGGTCTTATTCCAAGAGTAGGAGATCTAGGTCCTACCACGACCGCTCCAGTTCTCGGTCGCGGAAGCGTTCCAAGTCACGGAGCAGATCCAGAACCCCTGCATCAAAACGTTCGACGAGAGGTACCGGTGGAGAGGGAAGAAGTCCTCCAGTCCAGAATATCGACTCCAGGCAGCGTGTGTCGCCTGTGCCTCAGGAGGCGACGAATGGTCCGACCGACGGCTGGTCACCTTCCAAAGGCGATGAGAGATGGTCTTCCAGTCCCAAGGAGCAGTGA
Protein
MIQPESFSLLKVSKHSSEVIRFDAEIENYSMLKMVIMRLDERVVKLNDYPEPLRVKVCEAKSDFPSRHAWDSFFRDASDMDEMKPGERPDTIHISNLPIRWFVHKNERDEDAMPSESLFKKVFEKFGRVRQVDIPIADPFRLQMKSAIRGIMYPTEDAAVYFEGYVQFNEYVGFVRCMDELRGKKLVRKQDDIAEWCSIRVDFDKTKHMTDAAIKRRAIVRERLAARKSHKEEEEQAEKLKAAKREAKERQKYELAEKEKLEKMRQREERRKKKQLAKLMERDNVDLNSKVAEEQKKLLKAQKKLQAIRLVAELFKRIELRPELQRNGAKTHRAYNESLRARVVEKFKRQQEELLEAQRKNLRSALDGRIVIRSALETKKPRRTSSLSSLSADEENQLRIKTERIPSPDSRMQYDKTPVPAMYGMANPYSFGYPAFMPPGYAYGPQSSMYYPRAAYPAEWARRAGGARGRGRGVRGRGGARPRMPYFDGPDATAQYYRYFKKLKESNEHDDRLHSRSRSRSRSRSRRRSYTRSRSRRRSYSRSRSRSRRSRSRSRSRSRSKSRSRRSYSKSRRSRSYHDRSSSRSRKRSKSRSRSRTPASKRSTRGTGGEGRSPPVQNIDSRQRVSPVPQEATNGPTDGWSPSKGDERWSSSPKEQ
Summary
Uniprot
EMBL
Proteomes
Pfam
PF00089 Trypsin
Interpro
Gene 3D
ProteinModelPortal
Ontologies
KEGG
GO
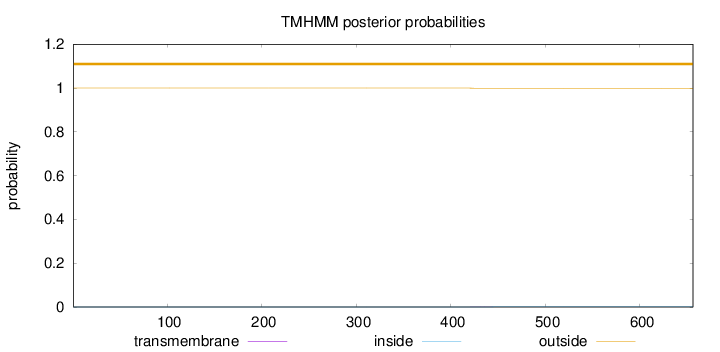
Topology
Length:
656
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01551
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00006
outside
1 - 656
Population Genetic Test Statistics
Pi
18.894012
Theta
24.195104
Tajima's D
-1.051404
CLR
138.837154
CSRT
0.122793860306985
Interpretation
Possibly Positive selection
