Gene
KWMTBOMO12723 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007183
Annotation
serine_protease_38_[Mamestra_configurata]
Location in the cell
Extracellular Reliability : 1.007
Sequence
CDS
ATGAAGGTTCTATCTTTTGTACTATTGTGCGGTCTTGCTCTGGTACAGGGGCGCAGCACCGGTGTGCAAAGTACCAACGCCCTGGTTGCCCTCGGAGACAAGCCCTACCAGGTCCATCTACGCATCGCTGTTTCGACCAGTGGTCTGCTCAACACCTGCGCCGGTTCCCTCATTCACTCTCGCTGGGTCCTCAGCGCCGCTAGCTGCCTCCAAGATGTCCGTTTCATCTGGGTCCGTTATGGTCTGGTGGTGGTAATTAACCCGTCCCTGGTCACGGAGACTAGCGCAGTCCGTCTGCACCCCTCGGATACCATTGGTCTCGTCAGCATCAACAGGGATGTCCAACCCACTGACTTCATCTCTCCCGTGGCTTTGTCTGCCAGCGAGGACCTACCCGAATCCGGAAATGTCTGCGGCTTTGGCGAAGTCGACGGCGAACCTGGAGAGCAACTGAGCTGCTTCGACGTGTCCGTGGTGCCCGCCGACGGTCTCCTTGAGGCCACCAGCGAGGAAGGCCAGACTTCCAAGTACGATGTTGGAACTGCTCTTGTCAGCGATGATGTTCAAGTAGCTGTGCTCCTGGCCGGTGCTGACGAGAACAGCGCTGGAACCTTCGTGCCTGTCGCGGAATACATCGAATGGATCGAGACCACCGCCGGTATTACCCTGGCTCCTGAAGAACCTGAGTCTTCCGAATCTTCCTCATCTTCTTCATCTTCTGAGGAAGACGAAGAAGAAGAAGAGGAAGAAGAAGAGGAAGAGGAAGAAGAAGAGTAA
Protein
MKVLSFVLLCGLALVQGRSTGVQSTNALVALGDKPYQVHLRIAVSTSGLLNTCAGSLIHSRWVLSAASCLQDVRFIWVRYGLVVVINPSLVTETSAVRLHPSDTIGLVSINRDVQPTDFISPVALSASEDLPESGNVCGFGEVDGEPGEQLSCFDVSVVPADGLLEATSEEGQTSKYDVGTALVSDDVQVAVLLAGADENSAGTFVPVAEYIEWIETTAGITLAPEEPESSESSSSSSSSEEDEEEEEEEEEEEEEEE
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JCD8
C9W8I0
A0A1E1VYP9
A0A125QVZ5
A0A2H1WDZ4
B4X993
+ More
A9P3S6 O18446 O18449 A0A1B0RHN0 A0A2W1CP94 A0A0N1IP01 B6D1R2 A0A212F9H9 A0A0N1PFJ0 A0A452E0M8 A0A2K5EIC3 A0A3B3U7R5 A0A3P9J8D6 W5PFX3 A0A3P9LES0 A0A452EI21 A0A3B3ICG1 A0A3P9ML71 A0A383ZNL5 A0A3B3HJ78 A0A2K5SI43 A0A3P9MLG3 A0A3P9J949 A0A3P9LEC1 A0A3B3INT4 A0A3P9L0G8 A0A093J9C7 A0A3P9HR28 A0A3P9LF62 A0A1W4X025 H2L6Z3 A0A3P9LEC7 A0A3B3YYN3 L8ISS6 A0A3B3UYY4 A0A3B3I9T3 A0A3P9HRN1 A0A3B3VQ55 A0A3P8P0G2 A0A3B3HIB7 A0A3P9MCH8 A0A060XIV3 A0A3B3U9G1 A0A3P9HRL3 A0A3P9C721 A0A2U9B0F2 A0A3B3XQU4 A0A232EZ15 A0A437D5J9 A0A3P8VKH3 A0A3P9J8F8 A0A3P8VJW7 A0A443SDQ2 A0A3P8QBN9 A0A3P9MM49 A0A3B3VJW2 A0A3P9L0K9 A0A3B3IBY7 A0A3B3HP20 A0A3P9L2C4 A0A3B3UHX3 A0A3B3VQN8 A0A3B3VQS6 A0A3B3IHH1 A0A3P8QCU5 A0A3B3BL71
A9P3S6 O18446 O18449 A0A1B0RHN0 A0A2W1CP94 A0A0N1IP01 B6D1R2 A0A212F9H9 A0A0N1PFJ0 A0A452E0M8 A0A2K5EIC3 A0A3B3U7R5 A0A3P9J8D6 W5PFX3 A0A3P9LES0 A0A452EI21 A0A3B3ICG1 A0A3P9ML71 A0A383ZNL5 A0A3B3HJ78 A0A2K5SI43 A0A3P9MLG3 A0A3P9J949 A0A3P9LEC1 A0A3B3INT4 A0A3P9L0G8 A0A093J9C7 A0A3P9HR28 A0A3P9LF62 A0A1W4X025 H2L6Z3 A0A3P9LEC7 A0A3B3YYN3 L8ISS6 A0A3B3UYY4 A0A3B3I9T3 A0A3P9HRN1 A0A3B3VQ55 A0A3P8P0G2 A0A3B3HIB7 A0A3P9MCH8 A0A060XIV3 A0A3B3U9G1 A0A3P9HRL3 A0A3P9C721 A0A2U9B0F2 A0A3B3XQU4 A0A232EZ15 A0A437D5J9 A0A3P8VKH3 A0A3P9J8F8 A0A3P8VJW7 A0A443SDQ2 A0A3P8QBN9 A0A3P9MM49 A0A3B3VJW2 A0A3P9L0K9 A0A3B3IBY7 A0A3B3HP20 A0A3P9L2C4 A0A3B3UHX3 A0A3B3VQN8 A0A3B3VQS6 A0A3B3IHH1 A0A3P8QCU5 A0A3B3BL71
Pubmed
EMBL
BABH01031528
BABH01031529
FJ205426
ACR15993.2
GDQN01011191
JAT79863.1
+ More
KP690085 ALO61087.1 ODYU01008026 SOQ51289.1 EF531629 ABR88240.1 EF104915 ABP96917.1 Y12282 CAA72961.1 Y12286 CAA72965.1 KM360187 AKH49603.1 NHMN01024378 PZC87414.1 KQ460882 KPJ11503.1 EU673455 ACI45418.1 AGBW02009568 OWR50405.1 KPJ11502.1 LWLT01000008 AMGL01092701 KL217258 KFV75544.1 JH880825 ELR58524.1 FR905449 CDQ79391.1 CP026244 AWO97393.1 NNAY01001573 OXU23549.1 CM012444 RVE69987.1 NCKV01003494 RWS25653.1
KP690085 ALO61087.1 ODYU01008026 SOQ51289.1 EF531629 ABR88240.1 EF104915 ABP96917.1 Y12282 CAA72961.1 Y12286 CAA72965.1 KM360187 AKH49603.1 NHMN01024378 PZC87414.1 KQ460882 KPJ11503.1 EU673455 ACI45418.1 AGBW02009568 OWR50405.1 KPJ11502.1 LWLT01000008 AMGL01092701 KL217258 KFV75544.1 JH880825 ELR58524.1 FR905449 CDQ79391.1 CP026244 AWO97393.1 NNAY01001573 OXU23549.1 CM012444 RVE69987.1 NCKV01003494 RWS25653.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JCD8
C9W8I0
A0A1E1VYP9
A0A125QVZ5
A0A2H1WDZ4
B4X993
+ More
A9P3S6 O18446 O18449 A0A1B0RHN0 A0A2W1CP94 A0A0N1IP01 B6D1R2 A0A212F9H9 A0A0N1PFJ0 A0A452E0M8 A0A2K5EIC3 A0A3B3U7R5 A0A3P9J8D6 W5PFX3 A0A3P9LES0 A0A452EI21 A0A3B3ICG1 A0A3P9ML71 A0A383ZNL5 A0A3B3HJ78 A0A2K5SI43 A0A3P9MLG3 A0A3P9J949 A0A3P9LEC1 A0A3B3INT4 A0A3P9L0G8 A0A093J9C7 A0A3P9HR28 A0A3P9LF62 A0A1W4X025 H2L6Z3 A0A3P9LEC7 A0A3B3YYN3 L8ISS6 A0A3B3UYY4 A0A3B3I9T3 A0A3P9HRN1 A0A3B3VQ55 A0A3P8P0G2 A0A3B3HIB7 A0A3P9MCH8 A0A060XIV3 A0A3B3U9G1 A0A3P9HRL3 A0A3P9C721 A0A2U9B0F2 A0A3B3XQU4 A0A232EZ15 A0A437D5J9 A0A3P8VKH3 A0A3P9J8F8 A0A3P8VJW7 A0A443SDQ2 A0A3P8QBN9 A0A3P9MM49 A0A3B3VJW2 A0A3P9L0K9 A0A3B3IBY7 A0A3B3HP20 A0A3P9L2C4 A0A3B3UHX3 A0A3B3VQN8 A0A3B3VQS6 A0A3B3IHH1 A0A3P8QCU5 A0A3B3BL71
A9P3S6 O18446 O18449 A0A1B0RHN0 A0A2W1CP94 A0A0N1IP01 B6D1R2 A0A212F9H9 A0A0N1PFJ0 A0A452E0M8 A0A2K5EIC3 A0A3B3U7R5 A0A3P9J8D6 W5PFX3 A0A3P9LES0 A0A452EI21 A0A3B3ICG1 A0A3P9ML71 A0A383ZNL5 A0A3B3HJ78 A0A2K5SI43 A0A3P9MLG3 A0A3P9J949 A0A3P9LEC1 A0A3B3INT4 A0A3P9L0G8 A0A093J9C7 A0A3P9HR28 A0A3P9LF62 A0A1W4X025 H2L6Z3 A0A3P9LEC7 A0A3B3YYN3 L8ISS6 A0A3B3UYY4 A0A3B3I9T3 A0A3P9HRN1 A0A3B3VQ55 A0A3P8P0G2 A0A3B3HIB7 A0A3P9MCH8 A0A060XIV3 A0A3B3U9G1 A0A3P9HRL3 A0A3P9C721 A0A2U9B0F2 A0A3B3XQU4 A0A232EZ15 A0A437D5J9 A0A3P8VKH3 A0A3P9J8F8 A0A3P8VJW7 A0A443SDQ2 A0A3P8QBN9 A0A3P9MM49 A0A3B3VJW2 A0A3P9L0K9 A0A3B3IBY7 A0A3B3HP20 A0A3P9L2C4 A0A3B3UHX3 A0A3B3VQN8 A0A3B3VQS6 A0A3B3IHH1 A0A3P8QCU5 A0A3B3BL71
PDB
2ZGJ
E-value=5.7183e-07,
Score=126
Ontologies
GO
PANTHER
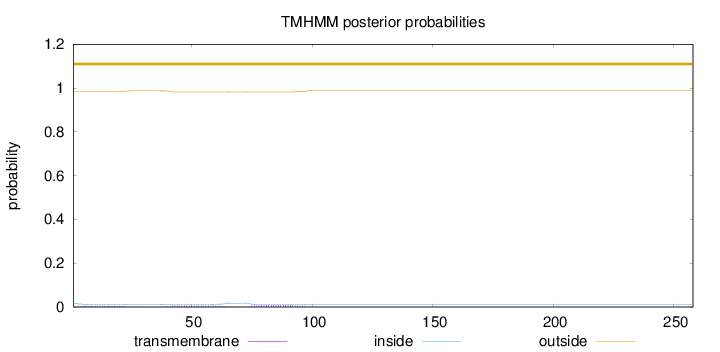
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.995218
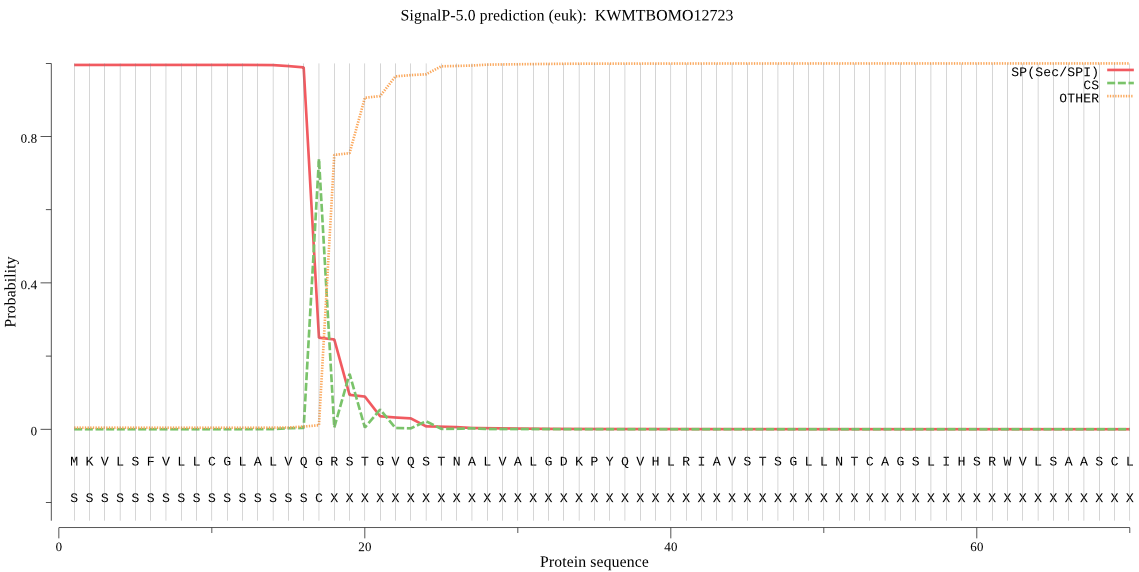
Length:
258
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.43342
Exp number, first 60 AAs:
0.23607
Total prob of N-in:
0.01690
outside
1 - 258
Population Genetic Test Statistics
Pi
0.804653
Theta
4.599658
Tajima's D
-2.033569
CLR
153.354961
CSRT
0.00874956252187391
Interpretation
Possibly Positive selection
