Gene
KWMTBOMO12722
Pre Gene Modal
BGIBMGA007184
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_cytochrome_P450_6B1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.24
Sequence
CDS
ATGTTTGGCTTCCGGAGATCATTGGTTGAAGCCGTGGTGTTCCTCTCGTGGCTCTTAGAAGACTGGACATTCGGCTCCAGCGTCCTATGGCACGTGTATTTTACTCTCTGGATCTGCAGCCTCGTCCTGACCGTGACCACATACGATCATGACTACTGGAAGGACAGAGGCATATTCTCACCCCCGGCCTTGCCCGTGGTTGGACATATACTGTCTGTTGTCCTATTCAGGGAGCAGGGAGGGATGTGCTTCAAAAGGATCTATGATACGTATACAGATCAACGGTTTTTAGGTACGCATCAGTTCTATCAGCGCACGTTGGTAATCAGAGATCCAGAGCTCATTAAAAGGGTCTGCGTGAACGATTTCCAGCATTTCGTGGACCGTGGATTCTTCTTCAACAAGGAAGTAGATCCTTTGGCTGGATCGGTACTTTTCCTGCGAGGAAACGAATGGAAAAAATTAAGAGCCAAAATTTCGCCAATCTTTTCGCCGAACAAGCTACGAGGAATGTTCCCGTTGATAGACAACACAGCCGATGAGTTCGTCATACGTTTGAGGAAACGAATCAAAAACAAGAACAAGGATATCGAAAACGTGGACAGCAATTCTAATATAGAAGATGCTTATGCGCTGGTGGACTCTGAGGAGTTGGTTGGGGGGTATACAGCCGACGCGATAGTACCTTGCGCCTTTGGACTGAAGAGTCTAGTTATGGATAAGCCTGACGATCCATTTGCGGTTGCACTTCAAGCGTTTTACGAGAAATCTTTATATAATGTATTTGAAAAAACGATGCGTCAGTTCTGGCCAGCATTCGTCCTCTTCTTTAGAATGAGGATAATACCGAAGAAGACTCACGATTTCTTCCATAACGTCGTGACAAGCGTGCTCAGAGCTCGCGAAAGTGGTGAACAGGAGAAACGTGGTGACTTCATCGACATGATGATGAATCTACGAAACGATGACGCCAATAATAAAAAAACAGACGATCCTGATGAAGTAGAAATAACGGACATAGTAATCTCCGCCAACGCATTTATTATATTCCTTGGTGGATTTGAAACTACTTCTTCGACCTTAGCGTTTCTCTTCCTCGAACTGGCAGCTGATCAGAGGGTCCAGGAGAAGATGAGACAGGAGATTAGAGAGGTGTTGAGCCGAAATGACGGAAAGATGACTTTTGAGATCTTACAAGAGTTAATCTACATGGAGATGGTAATACAAGAGACCCTGCGTTTGTATCCACCCTTTCCAAGCATACAACGTATGTGTACCAAAGACTATCTTATACCGGGAACCGAGAAAGTCGTTGAGAAAGGCACAATAGTTCTGTTTCCGACGCTGGGAATCCAAAGGGACAACCAATACTTCGAACGCGCGGATGACTTTTATCCTGAGAGGTGGTCAGGAGGGTTCGTCCCTCCGCCAGGAGTCTATATGCCTTTTGGGGATGGACCTCGTTATTGTATAGGTAAAAGGTTCGCAATGATCCAGATGAAGTGCTGTCTAGTCAAAGTGTTGCAGTACGTCGTGATCACGCCGGCGTCTCCTGCTCAGCCGCGGACCAGACCCTTCGCGGCAGACTGTTGCTCTCCCCAAACCCTCCACCCCGACAACTCCTTAGTCAAACTGAGCCTGTTATGA
Protein
MFGFRRSLVEAVVFLSWLLEDWTFGSSVLWHVYFTLWICSLVLTVTTYDHDYWKDRGIFSPPALPVVGHILSVVLFREQGGMCFKRIYDTYTDQRFLGTHQFYQRTLVIRDPELIKRVCVNDFQHFVDRGFFFNKEVDPLAGSVLFLRGNEWKKLRAKISPIFSPNKLRGMFPLIDNTADEFVIRLRKRIKNKNKDIENVDSNSNIEDAYALVDSEELVGGYTADAIVPCAFGLKSLVMDKPDDPFAVALQAFYEKSLYNVFEKTMRQFWPAFVLFFRMRIIPKKTHDFFHNVVTSVLRARESGEQEKRGDFIDMMMNLRNDDANNKKTDDPDEVEITDIVISANAFIIFLGGFETTSSTLAFLFLELAADQRVQEKMRQEIREVLSRNDGKMTFEILQELIYMEMVIQETLRLYPPFPSIQRMCTKDYLIPGTEKVVEKGTIVLFPTLGIQRDNQYFERADDFYPERWSGGFVPPPGVYMPFGDGPRYCIGKRFAMIQMKCCLVKVLQYVVITPASPAQPRTRPFAADCCSPQTLHPDNSLVKLSLL
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
A0A2W1BA48
H9JCD9
A0A194QG23
A0A0N1IG98
A0A437BE96
A0A2A4J6C1
+ More
A0A146LS76 A0A0A9WUZ2 A0A1B6CVZ0 A0A1B6CZQ5 A0A1Q1NKX5 A0A0K0LBA0 J9K291 A0A2D1GSE2 A0A2L1K3L2 A0A2S2NDH0 K4K2T0 A0A2H8TVC3 A0A165RB50 A0A0K1YW70 A0A2J7PVE9 A0A2J7PVE5 R9UDU6 A0A2S2Q9N3 M9TLT7 A0A2J7R4P3 A0A2J7QXU7 A0A2J7R4Q0 A0A2J7R4P8 A0A2J7R4Q3 D6WB59 A0A2J7PVE4 A0A1B6EZI3 A0A1B6L6W5 B6E554 A0A1B6FAE0 A0A288CXN3 A0A1B6FYS7 J9JKC4 Q5W1K7 D2A0A3 A0A0M8ZWI2 J9LU05 A0A2J7PVD8 W6E8Q3 R4WT71 A0A0L7QLZ6 A0A232F263 A0A2D1GSD1 K7J4D7 X1XTT6 A0A0L7LIR5 A0A2J7QIZ0 J9K0E4 A0A0K8TV67 J9JTS8 J9JXE5 I4DJQ1 A0A182UMK3 J9JLB8 A0A1B6K0J3 B4J6T0 A0A1W4WJ93 A0A2S2PZX4 A0A084VFX6 A0A0A9VTP3 A0A1Q1NKX7 A0A1L2CDK3 A0A1L2CDV8 A0A1L2CDT2 A0A0A9VTP6 A0A026W444 A0A2P0QDL0 A0A1B6CGE4 A0A1L2CDP8 A0A1W4WLU0 A0A1L2CDR8 A0A194QG84 A0A2P0QFV8 A0A1L2CDP1 A0A1L1VD15 A0A146LKG2 A0A1B6KYX2 A0A1L2CDL4 A0A1L2CDM7 A0A1L2CDV1 A0A2Y9D1F7 A0A1B6KJS6 A0A340TB86 A0A2J7QIY4 A0A2D1GSF0 A0A2Y9D1S5 Q7Q449 A0A0T6B6B5 A0A182U2R2
A0A146LS76 A0A0A9WUZ2 A0A1B6CVZ0 A0A1B6CZQ5 A0A1Q1NKX5 A0A0K0LBA0 J9K291 A0A2D1GSE2 A0A2L1K3L2 A0A2S2NDH0 K4K2T0 A0A2H8TVC3 A0A165RB50 A0A0K1YW70 A0A2J7PVE9 A0A2J7PVE5 R9UDU6 A0A2S2Q9N3 M9TLT7 A0A2J7R4P3 A0A2J7QXU7 A0A2J7R4Q0 A0A2J7R4P8 A0A2J7R4Q3 D6WB59 A0A2J7PVE4 A0A1B6EZI3 A0A1B6L6W5 B6E554 A0A1B6FAE0 A0A288CXN3 A0A1B6FYS7 J9JKC4 Q5W1K7 D2A0A3 A0A0M8ZWI2 J9LU05 A0A2J7PVD8 W6E8Q3 R4WT71 A0A0L7QLZ6 A0A232F263 A0A2D1GSD1 K7J4D7 X1XTT6 A0A0L7LIR5 A0A2J7QIZ0 J9K0E4 A0A0K8TV67 J9JTS8 J9JXE5 I4DJQ1 A0A182UMK3 J9JLB8 A0A1B6K0J3 B4J6T0 A0A1W4WJ93 A0A2S2PZX4 A0A084VFX6 A0A0A9VTP3 A0A1Q1NKX7 A0A1L2CDK3 A0A1L2CDV8 A0A1L2CDT2 A0A0A9VTP6 A0A026W444 A0A2P0QDL0 A0A1B6CGE4 A0A1L2CDP8 A0A1W4WLU0 A0A1L2CDR8 A0A194QG84 A0A2P0QFV8 A0A1L2CDP1 A0A1L1VD15 A0A146LKG2 A0A1B6KYX2 A0A1L2CDL4 A0A1L2CDM7 A0A1L2CDV1 A0A2Y9D1F7 A0A1B6KJS6 A0A340TB86 A0A2J7QIY4 A0A2D1GSF0 A0A2Y9D1S5 Q7Q449 A0A0T6B6B5 A0A182U2R2
Pubmed
EMBL
KZ150436
PZC70874.1
BABH01031525
BABH01031526
KQ458981
KPJ04498.1
+ More
KQ460882 KPJ11504.1 RSAL01000080 RVE48658.1 NWSH01002695 PCG67667.1 GDHC01009339 JAQ09290.1 GBHO01033271 JAG10333.1 GEDC01019701 JAS17597.1 GEDC01018434 JAS18864.1 KX660704 AQM57043.1 KM217019 AIW79982.1 ABLF02039155 MF471404 ATN96010.1 MG595274 AVE15906.1 GGMR01002624 MBY15243.1 JX566819 AFU86482.1 GFXV01005707 MBW17512.1 KR028424 MF471403 AML23850.1 ATN96009.1 KP859386 AKZ17696.1 NEVH01020964 PNF20301.1 PNF20303.1 JX462997 AGN52790.1 GGMS01005256 MBY74459.1 KC683904 AGJ51945.1 NEVH01007403 PNF35808.1 NEVH01009372 PNF33413.1 PNF35811.1 PNF35807.1 PNF35809.1 KQ971311 EEZ99338.1 PNF20299.1 GECZ01026392 GECZ01007074 JAS43377.1 JAS62695.1 GEBQ01020616 JAT19361.1 FJ209361 ACI25098.1 GECZ01022886 JAS46883.1 JN984153 AFR43484.1 GECZ01014556 GECZ01011176 JAS55213.1 JAS58593.1 ABLF02040587 AJ852423 CAH65682.2 KQ971338 EFA02817.1 KQ435817 KOX72550.1 ABLF02031675 PNF20302.1 KF896073 AHJ10931.1 AK417927 BAN21142.1 KQ414906 KOC59584.1 NNAY01001191 OXU24791.1 MF471387 ATN95993.1 JTDY01001023 KOB75101.1 NEVH01013565 PNF28550.1 GCVX01000074 JAI18156.1 ABLF02040873 ABLF02040879 ABLF02040890 ABLF02040579 ABLF02040580 ABLF02040583 ABLF02042802 AK401519 BAM18141.1 ABLF02026945 GECU01002739 JAT04968.1 CH916367 EDW02011.1 GGMS01001249 MBY70452.1 ATLV01012547 KE524804 KFB36870.1 GBHO01043957 JAF99646.1 KX660703 AQM57042.1 KU169066 KU169067 AMH87378.1 AMH87379.1 KU169082 KU169095 KU169100 KU169101 AMH87394.1 AMH87407.1 AMH87412.1 AMH87413.1 KU169115 KU169121 AMH87427.1 AMH87433.1 GBHO01043952 JAF99651.1 KK107447 QOIP01000001 EZA50778.1 RLU27579.1 KY584290 KY584292 ARM59292.1 ARM59294.1 GEDC01024878 GEDC01019823 JAS12420.1 JAS17475.1 KU169079 KU169083 AMH87391.1 AMH87395.1 KU169104 AMH87416.1 KPJ04502.1 KY584294 KY584295 ARM59295.1 ARM59296.1 KU169081 AMH87393.1 JN984869 AFU75895.1 GDHC01010046 JAQ08583.1 GEBQ01031744 GEBQ01023329 GEBQ01015122 GEBQ01014486 GEBQ01005724 JAT08233.1 JAT16648.1 JAT24855.1 JAT25491.1 JAT34253.1 KU169000 KU169001 KU169002 KU169003 KU169004 KU169010 KU169011 AMH87312.1 AMH87313.1 AMH87314.1 AMH87315.1 AMH87316.1 AMH87322.1 AMH87323.1 KU169058 KU169059 AMH87370.1 AMH87371.1 KU169090 AMH87402.1 GEBQ01028287 GEBQ01007193 JAT11690.1 JAT32784.1 PNF28547.1 MF471395 ATN96001.1 AAAB01008964 EAA12401.2 LJIG01009517 KRT82922.1
KQ460882 KPJ11504.1 RSAL01000080 RVE48658.1 NWSH01002695 PCG67667.1 GDHC01009339 JAQ09290.1 GBHO01033271 JAG10333.1 GEDC01019701 JAS17597.1 GEDC01018434 JAS18864.1 KX660704 AQM57043.1 KM217019 AIW79982.1 ABLF02039155 MF471404 ATN96010.1 MG595274 AVE15906.1 GGMR01002624 MBY15243.1 JX566819 AFU86482.1 GFXV01005707 MBW17512.1 KR028424 MF471403 AML23850.1 ATN96009.1 KP859386 AKZ17696.1 NEVH01020964 PNF20301.1 PNF20303.1 JX462997 AGN52790.1 GGMS01005256 MBY74459.1 KC683904 AGJ51945.1 NEVH01007403 PNF35808.1 NEVH01009372 PNF33413.1 PNF35811.1 PNF35807.1 PNF35809.1 KQ971311 EEZ99338.1 PNF20299.1 GECZ01026392 GECZ01007074 JAS43377.1 JAS62695.1 GEBQ01020616 JAT19361.1 FJ209361 ACI25098.1 GECZ01022886 JAS46883.1 JN984153 AFR43484.1 GECZ01014556 GECZ01011176 JAS55213.1 JAS58593.1 ABLF02040587 AJ852423 CAH65682.2 KQ971338 EFA02817.1 KQ435817 KOX72550.1 ABLF02031675 PNF20302.1 KF896073 AHJ10931.1 AK417927 BAN21142.1 KQ414906 KOC59584.1 NNAY01001191 OXU24791.1 MF471387 ATN95993.1 JTDY01001023 KOB75101.1 NEVH01013565 PNF28550.1 GCVX01000074 JAI18156.1 ABLF02040873 ABLF02040879 ABLF02040890 ABLF02040579 ABLF02040580 ABLF02040583 ABLF02042802 AK401519 BAM18141.1 ABLF02026945 GECU01002739 JAT04968.1 CH916367 EDW02011.1 GGMS01001249 MBY70452.1 ATLV01012547 KE524804 KFB36870.1 GBHO01043957 JAF99646.1 KX660703 AQM57042.1 KU169066 KU169067 AMH87378.1 AMH87379.1 KU169082 KU169095 KU169100 KU169101 AMH87394.1 AMH87407.1 AMH87412.1 AMH87413.1 KU169115 KU169121 AMH87427.1 AMH87433.1 GBHO01043952 JAF99651.1 KK107447 QOIP01000001 EZA50778.1 RLU27579.1 KY584290 KY584292 ARM59292.1 ARM59294.1 GEDC01024878 GEDC01019823 JAS12420.1 JAS17475.1 KU169079 KU169083 AMH87391.1 AMH87395.1 KU169104 AMH87416.1 KPJ04502.1 KY584294 KY584295 ARM59295.1 ARM59296.1 KU169081 AMH87393.1 JN984869 AFU75895.1 GDHC01010046 JAQ08583.1 GEBQ01031744 GEBQ01023329 GEBQ01015122 GEBQ01014486 GEBQ01005724 JAT08233.1 JAT16648.1 JAT24855.1 JAT25491.1 JAT34253.1 KU169000 KU169001 KU169002 KU169003 KU169004 KU169010 KU169011 AMH87312.1 AMH87313.1 AMH87314.1 AMH87315.1 AMH87316.1 AMH87322.1 AMH87323.1 KU169058 KU169059 AMH87370.1 AMH87371.1 KU169090 AMH87402.1 GEBQ01028287 GEBQ01007193 JAT11690.1 JAT32784.1 PNF28547.1 MF471395 ATN96001.1 AAAB01008964 EAA12401.2 LJIG01009517 KRT82922.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BA48
H9JCD9
A0A194QG23
A0A0N1IG98
A0A437BE96
A0A2A4J6C1
+ More
A0A146LS76 A0A0A9WUZ2 A0A1B6CVZ0 A0A1B6CZQ5 A0A1Q1NKX5 A0A0K0LBA0 J9K291 A0A2D1GSE2 A0A2L1K3L2 A0A2S2NDH0 K4K2T0 A0A2H8TVC3 A0A165RB50 A0A0K1YW70 A0A2J7PVE9 A0A2J7PVE5 R9UDU6 A0A2S2Q9N3 M9TLT7 A0A2J7R4P3 A0A2J7QXU7 A0A2J7R4Q0 A0A2J7R4P8 A0A2J7R4Q3 D6WB59 A0A2J7PVE4 A0A1B6EZI3 A0A1B6L6W5 B6E554 A0A1B6FAE0 A0A288CXN3 A0A1B6FYS7 J9JKC4 Q5W1K7 D2A0A3 A0A0M8ZWI2 J9LU05 A0A2J7PVD8 W6E8Q3 R4WT71 A0A0L7QLZ6 A0A232F263 A0A2D1GSD1 K7J4D7 X1XTT6 A0A0L7LIR5 A0A2J7QIZ0 J9K0E4 A0A0K8TV67 J9JTS8 J9JXE5 I4DJQ1 A0A182UMK3 J9JLB8 A0A1B6K0J3 B4J6T0 A0A1W4WJ93 A0A2S2PZX4 A0A084VFX6 A0A0A9VTP3 A0A1Q1NKX7 A0A1L2CDK3 A0A1L2CDV8 A0A1L2CDT2 A0A0A9VTP6 A0A026W444 A0A2P0QDL0 A0A1B6CGE4 A0A1L2CDP8 A0A1W4WLU0 A0A1L2CDR8 A0A194QG84 A0A2P0QFV8 A0A1L2CDP1 A0A1L1VD15 A0A146LKG2 A0A1B6KYX2 A0A1L2CDL4 A0A1L2CDM7 A0A1L2CDV1 A0A2Y9D1F7 A0A1B6KJS6 A0A340TB86 A0A2J7QIY4 A0A2D1GSF0 A0A2Y9D1S5 Q7Q449 A0A0T6B6B5 A0A182U2R2
A0A146LS76 A0A0A9WUZ2 A0A1B6CVZ0 A0A1B6CZQ5 A0A1Q1NKX5 A0A0K0LBA0 J9K291 A0A2D1GSE2 A0A2L1K3L2 A0A2S2NDH0 K4K2T0 A0A2H8TVC3 A0A165RB50 A0A0K1YW70 A0A2J7PVE9 A0A2J7PVE5 R9UDU6 A0A2S2Q9N3 M9TLT7 A0A2J7R4P3 A0A2J7QXU7 A0A2J7R4Q0 A0A2J7R4P8 A0A2J7R4Q3 D6WB59 A0A2J7PVE4 A0A1B6EZI3 A0A1B6L6W5 B6E554 A0A1B6FAE0 A0A288CXN3 A0A1B6FYS7 J9JKC4 Q5W1K7 D2A0A3 A0A0M8ZWI2 J9LU05 A0A2J7PVD8 W6E8Q3 R4WT71 A0A0L7QLZ6 A0A232F263 A0A2D1GSD1 K7J4D7 X1XTT6 A0A0L7LIR5 A0A2J7QIZ0 J9K0E4 A0A0K8TV67 J9JTS8 J9JXE5 I4DJQ1 A0A182UMK3 J9JLB8 A0A1B6K0J3 B4J6T0 A0A1W4WJ93 A0A2S2PZX4 A0A084VFX6 A0A0A9VTP3 A0A1Q1NKX7 A0A1L2CDK3 A0A1L2CDV8 A0A1L2CDT2 A0A0A9VTP6 A0A026W444 A0A2P0QDL0 A0A1B6CGE4 A0A1L2CDP8 A0A1W4WLU0 A0A1L2CDR8 A0A194QG84 A0A2P0QFV8 A0A1L2CDP1 A0A1L1VD15 A0A146LKG2 A0A1B6KYX2 A0A1L2CDL4 A0A1L2CDM7 A0A1L2CDV1 A0A2Y9D1F7 A0A1B6KJS6 A0A340TB86 A0A2J7QIY4 A0A2D1GSF0 A0A2Y9D1S5 Q7Q449 A0A0T6B6B5 A0A182U2R2
PDB
6MA7
E-value=4.85819e-54,
Score=535
Ontologies
GO
Topology
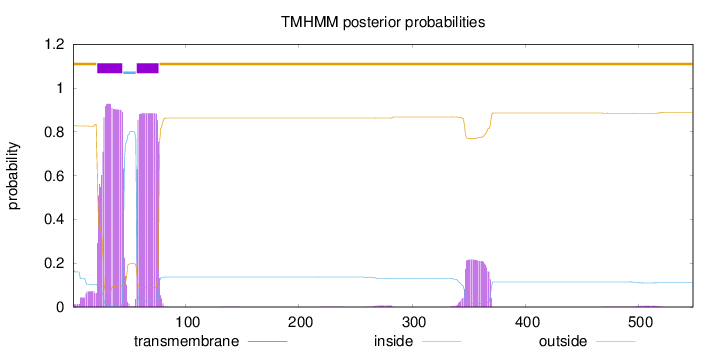
Length:
548
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.4275300000001
Exp number, first 60 AAs:
24.03812
Total prob of N-in:
0.17247
POSSIBLE N-term signal
sequence
outside
1 - 21
TMhelix
22 - 44
inside
45 - 56
TMhelix
57 - 76
outside
77 - 548
Population Genetic Test Statistics
Pi
23.480761
Theta
19.292354
Tajima's D
-1.45339
CLR
1.052626
CSRT
0.0658467076646168
Interpretation
Uncertain
