Gene
KWMTBOMO12719
Pre Gene Modal
BGIBMGA014022
Annotation
PREDICTED:_collagenase-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 2.83
Sequence
CDS
ATGCGTTATTCTGTCTTATTCATACTGACATGTGCAGTTATATTTGCGAATGGGTATGCTTTCAATGGACCTGCATACGGTTATCATTTGAGGTTCGGAGTGCCCAAAGCATATGAGTTGAAGCGTTTGGAGTCCAGTAGGATTATTGGAGGAAGCCAAGTTGCAGCGGCATCTGTGATACCACATCATGTAGGACTAGTGGCGCTCTTAACGACCGGTTGGATGTCAATCTGCGGTGGCTCTCTTGTCTCCAACACGAGGATCATCACTGCAGCTCACTGCTGGTGGGACGGCCAGAGTCAAGCGAAGCAGTTCACAGCAGTACTCGGCTCCACGACTATATTCACTGGGGGGATCAGGATTGACACCTCAGCTGTAACTGTCCACCCTAATTGGAACACTAACGATTACACCCATGATATTGCCGTGATAAGGATCACTAGTGTTGCATACAGTAATAATGTTCAAGCGATTCCATTGCCATCGGCCAGCGATGTCAACCGCGACTTCGTAGGGTTGACTGGCACCATATCAGGATATGGAAAAACGAGTGGTGTGCTATACCAGACAAGTGTGCCTGTGATAACCAACGCAGTCTGCCAGAATAGTTTCCCCATCACGATCCAGGGTAGTCATCTTTGCACCAGCGGCGCTGGTAACCGTGGCACCTGTGACGGTGACTCTGGCGGGCCCTTGACCGTTGTCTGGAATAATTCAAGAATTTTGATAGGTGTTATTTCGTTCGGTCCTAACGAAGGGTGTGAGGCTGGGAAGCCATCTGTGTACACAAGAGTCACTGCGTTCTTAACTTGGATTAATGCGCATTTATTCGGTATCTCTAAAGCCAAAGATATCTGGAAAAGTGAATGGTTTGGAGCTCTGAGGATTGTCGGAGGAAGTCAGGTTACCGCAGCGACTTTGATACCGTATCAGGCTGGAATTATTGTGACTTTGCCAAAAAGTGATACATCGATTTGCGGTGGAGCAGTTCTATCTAACACCAGAATAATCACTGCTGCTCATTGTTGGTGGGACGGAGAGAGTCAGGCTACCACTTTTACTGCAGTCCTTGGATCTCTTACCATATTCTCAGGGGGAACAAGGATCGCGACTAAGGACGTGGTGACTCATTCATCATGGAACCCTCAGAATATTATAAATGATATTGCGATTGTGAAGATCCTTGCTGTAGTATTTAATAACAACATCCAAGCTGTTTCACTGCCCACTAATGCTCAGCAAGACTTTGCTGGTATGACAGCAACAGCTTCTGGATATGGAAAGAGAAGTGACTCTCAAGCATCTTTCCCTTCGTCAACATCCCTATACACTGTTTCACTAAGTATAATCACAAACAATCTGTGCCAGAGCAGTTTCAGTCTAAGGATTCAGAACAACCATATCTGCACCAGCGGCCAAGGTGGTAAAGGGACCTGTGATGGAGACTCAGGAGGACCTTTGACCGTTCTTACTAATAACCTAAAAACATTAGTGGGCATCGTGTCTTTTGGACTTGGCGATGGCTGTCAGTTGGGCTACCCCTCCGTCTTCACGCGTGTTACATCCTACTTAACTTGGATCAAAAGTAACATGTAA
Protein
MRYSVLFILTCAVIFANGYAFNGPAYGYHLRFGVPKAYELKRLESSRIIGGSQVAAASVIPHHVGLVALLTTGWMSICGGSLVSNTRIITAAHCWWDGQSQAKQFTAVLGSTTIFTGGIRIDTSAVTVHPNWNTNDYTHDIAVIRITSVAYSNNVQAIPLPSASDVNRDFVGLTGTISGYGKTSGVLYQTSVPVITNAVCQNSFPITIQGSHLCTSGAGNRGTCDGDSGGPLTVVWNNSRILIGVISFGPNEGCEAGKPSVYTRVTAFLTWINAHLFGISKAKDIWKSEWFGALRIVGGSQVTAATLIPYQAGIIVTLPKSDTSICGGAVLSNTRIITAAHCWWDGESQATTFTAVLGSLTIFSGGTRIATKDVVTHSSWNPQNIINDIAIVKILAVVFNNNIQAVSLPTNAQQDFAGMTATASGYGKRSDSQASFPSSTSLYTVSLSIITNNLCQSSFSLRIQNNHICTSGQGGKGTCDGDSGGPLTVLTNNLKTLVGIVSFGLGDGCQLGYPSVFTRVTSYLTWIKSNM
Summary
Similarity
Belongs to the peptidase S1 family.
Pubmed
EMBL
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
2HLC
E-value=9.63143e-31,
Score=334
Ontologies
GO
Topology
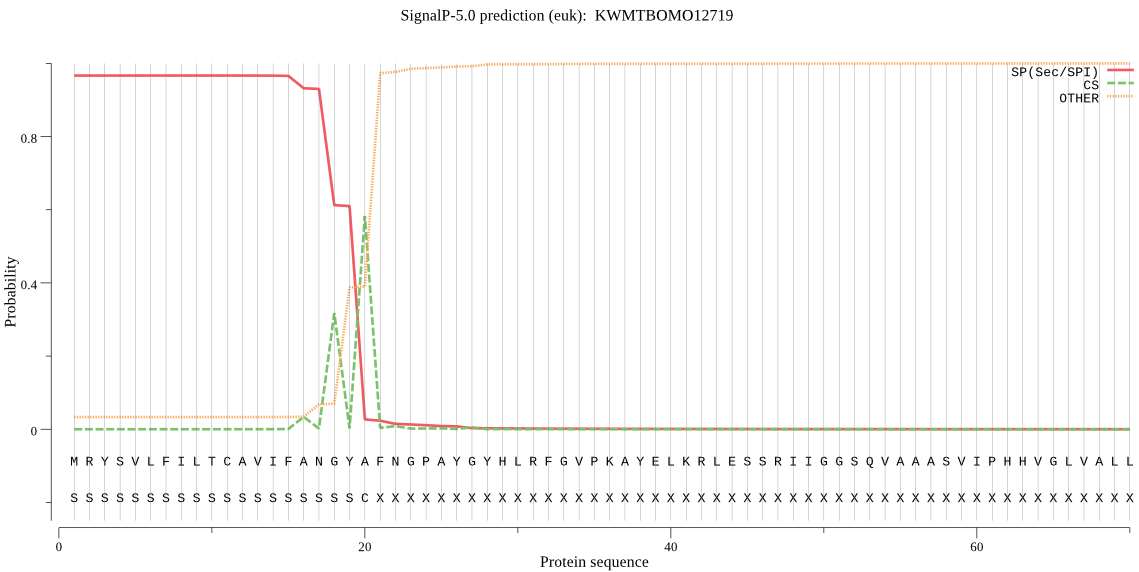
SignalP
Position: 1 - 20,
Likelihood: 0.965277
Length:
531
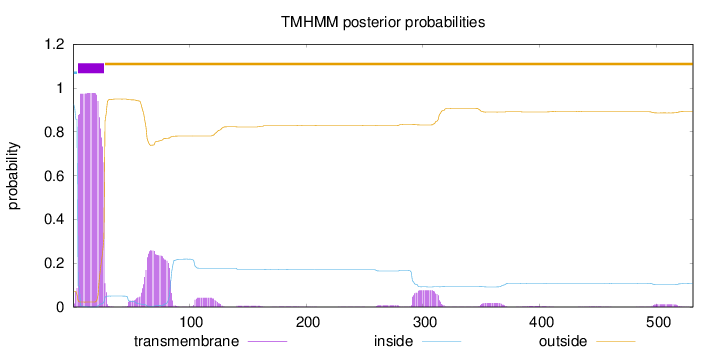
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
30.97967
Exp number, first 60 AAs:
22.07543
Total prob of N-in:
0.92776
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 531
Population Genetic Test Statistics
Pi
27.092229
Theta
24.153509
Tajima's D
-1.225549
CLR
14.419973
CSRT
0.0995950202489875
Interpretation
Uncertain
