Gene
KWMTBOMO12715
Pre Gene Modal
BGIBMGA013095
Annotation
reverse_transcriptase_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 3.154
Sequence
CDS
ATGGCTCAACTTATAGGAAACATATCTGAGTTTGATTATAAAACCCAAGAATGGCAGATCTTTTACAAGAAATTAGAAAATTTCCTAAAGTTGAACAAAGTTTTAGCTGAAGATTGGTGCTCCGTGCTTATTGCACATCTTTCAGATGATTCGTATCGGCTTGTGAGGAACCTCGTACATCCTAACGAGGTAGAAACAATACAATGGACTGAGCTGGTTAGAGTACTCAACGGACATTTTCAACCGAAAAGATCGACTTTTGCCGACAGGGCAAAATTTTATGGTGCGATCAGAATGGACGGTGAGAAAGTCGAAGATTGGGCGGCACGTCTGAGGGGACTCGCCGTGCACTGTAATTTCGGTACAGAATTAGATACGTTGCTACGTGATCGATTTATTCTAGGCCTGAAGGTAGGTCCAGAAAGAAATCGATTGTTCGAACAAGACTCGAACTCCCTAACGCTTTCAAGAGCTATTGAAATAGCCCAGGAGACTGCGTGTGCGCGGGCGGCGAGAGCTGATGTTGATGGTGGTGAGCAGCTTATCAAACAAGAGGTGGTGTATCGTGTCAGCGAGCGTCGGGCGCGGGGCGCCTCGACGCAGGCGGAGTCGTCGTGGGCATCGGCGGCAGCCGCGCGCTGTGCAGTGTGCGGGTTGAAAGGTCATTCCGTAGAAAAGTGCCGTTTCAAAGACTATAAGTGTATGATTTGTGGTAAAAAGGGTCACCTTAAGAAAGTGTGCAGTGTCAAAAAGGTCGGAAAACTGTCGTTAAATAATATGGATGATAACGAAAGCGATGGACACGAATGTAAAAAATGTGATTTGTATAATTTAAGGTATGTTCGCTACGATCCTATTAAGTTGAACATTTCTATAAATAATAAGTTTTATTCTATGGAATTAGATTGCGGAGCTGCTCGTAGTGTAATTTCAGAAAGATTTTACAGGGATAACTTCCCATTATACAAATTAAGCGCATCACCTGCAAGTATGATGTTTGGACGTAATTTAAGAACAAGATTAGATTTAATATTACCCACTAAAAAAGTGGAAGATAATGCAGTTGTTCCTAATAAAGAAATAAGTAAATCTAGACAATTCCAAATAGGTGAAATAGTCTGGGCAAGATTTTTTATTGCTCGAAAGGCTAGCTGGATGAAAGGAAAAATAACGAATATCTTAGGCAATAGAATGTTTCAGATCAATTTTGAAAACTGTGATTCAACGTGTGTCAGACATTTGGATCAGATTAGGAAGTACACGGATGCTAGGTCAAAAACAATTGTGAGCGATGTTGTTTCCACCGAAGGCGCTAATATTTCAGTCCCGGATCGACCCACAACAATGCAATCAGTATCGCAGCCTAGCTCTGAGATAGTGCTGTCCTCACCTATGTCGTCGCCGCCGGTACCGGAGCCGGCACCAGTATCGGCCCCCACGCACGGCAATAACGTTGACATTGTGAGGGGGGAGGATGAGGAGGGCGATGAATGGGAGGAGGCTAGTGAAAACATCGCAGCTGCCAGCCTGCCGACCGAGTGTGAACCTGATTACCCTATTGTAGCATCAGATGACCCCGATAACGTGAGATTAGAAGAGAGGGCGCCAGATAATGAAAGACAGTGTGGTCGACCAAAGAGGACAAGAAAAGTAGTAGATTATAAGAAGTATTTTTAA
Protein
MAQLIGNISEFDYKTQEWQIFYKKLENFLKLNKVLAEDWCSVLIAHLSDDSYRLVRNLVHPNEVETIQWTELVRVLNGHFQPKRSTFADRAKFYGAIRMDGEKVEDWAARLRGLAVHCNFGTELDTLLRDRFILGLKVGPERNRLFEQDSNSLTLSRAIEIAQETACARAARADVDGGEQLIKQEVVYRVSERRARGASTQAESSWASAAAARCAVCGLKGHSVEKCRFKDYKCMICGKKGHLKKVCSVKKVGKLSLNNMDDNESDGHECKKCDLYNLRYVRYDPIKLNISINNKFYSMELDCGAARSVISERFYRDNFPLYKLSASPASMMFGRNLRTRLDLILPTKKVEDNAVVPNKEISKSRQFQIGEIVWARFFIARKASWMKGKITNILGNRMFQINFENCDSTCVRHLDQIRKYTDARSKTIVSDVVSTEGANISVPDRPTTMQSVSQPSSEIVLSSPMSSPPVPEPAPVSAPTHGNNVDIVRGEDEEGDEWEEASENIAAASLPTECEPDYPIVASDDPDNVRLEERAPDNERQCGRPKRTRKVVDYKKYF
Summary
Uniprot
EMBL
Interpro
Gene 3D
ProteinModelPortal
Ontologies
KEGG
GO
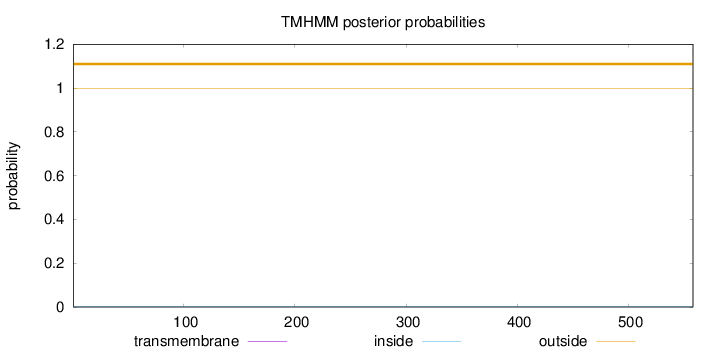
Topology
Length:
558
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000720000000000001
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00091
outside
1 - 558
Population Genetic Test Statistics
Pi
2.12112
Theta
21.232345
Tajima's D
-1.482704
CLR
45.875779
CSRT
0.0598470076496175
Interpretation
Uncertain
