Gene
KWMTBOMO12711
Pre Gene Modal
BGIBMGA001385
Annotation
PREDICTED:_spermine_oxidase-like_[Papilio_machaon]
Full name
Amine oxidase
Location in the cell
Cytoplasmic Reliability : 2.923
Sequence
CDS
ATGGACGTTATCGTAATAGGCTGCGGCGCGGCCGGTGTCGCAGCTTTAAGGAAACTCCATGACGCTGGTCTGCGAGTCCTCGGCTTGGAGGCAGCGGACAGAATCGGGGGTCGAATATGCACTATTGAATACGGTGACAGTACCCTGGATATCGGAGCTGCATGGTGTCATGGCGAAAAAGACAATATAGTGTTTGAACTAGCAGAGCCGCTAGGTCTACTTGGTAGACCAGACCCTCACGATAGCTGGTATGTACTATCCAATGGAGATCTGGCACCTGATGCGACATGCAAGGAAATACTTGCAAATATAGACGAAGAAGTATGTAAGAGTCACAAGAATAATGTACAGTCCATTTCGCAGTTTGTTAGGAATGCAGTAAACACTAAAGAAACATTTAAGCAATTTCCGAGATTAACCCGGAGTCTTTTAGAAGTATACGAAAGGAACAACCATTTAGGAGGTCAAGATGATCCTCAGCATGGCAAGAGCCTGAAGGGACTGGACGAACACTGGCCTTGTGAGGGGGAATTCTTGCTGAACTGGAGGGGGAGAGGTTATAAGACCTTACTGGACGTTTTGTTGAACAAATATCCAGACCCCAACGAAGCGATTCCCGTACAAATACTACTGAACAAACACGTGGAATGTATAAGATGGGGCACGAGCCAGCCTTCGCACCAAATAAGCCCGCTTGTGCAAGTGAAGTGTACCGATGGAAGCTTGTACGCTGCGAAAAGCGTTATTGTAACATTACCCTTAGCAGTACTGAAAGAGACACACGCTCAGCTCTTCAGCCCACCATTACCCCAAGACAAAATAAACTCAATCAACAGTTTGCATTATTGCGTGCTCGATAAAATCTACATAGAATTCACCACACCGTGGTGGCCAAAATCAGCCGGGAAATTCGTAATACTTTGGCAAGAAGAAGACAAGGCTAAATTCACCAAAGAAGAACATTGGATAACCGAAATATATGGCTTAGACCCTGTACAGCATCAGCCGAATGTGCTGCTTGCCTGGATTTATGGGAAAGGCGCTGAAGCTATGGAGAAAGTCAGCTTCGATGACCTGAAGGCTGGAATTGATAAATTACTGAGTATATTTAAGAAGAAATTCCCTGTTACCCCCGTTAAATCTGTTTTGAGGTCACAGTGGGCGTCAAATTTACTAGCGAGAAGCGCGTACGCGTACCGCTGTGTTGCCACTGAAGAAAACGGCGCTAGTGCGACAACACTGAGCGAGCCTATCTACCACGGCAATGGGTTGCCGTTAGTTTGCTTTGCCGGAGAAGCTACGTCGTATCACAGACACTCAGCTGTACATGGAGCGGTCGAGTCAGGATTCAGAGAGGCTCAACGACTTATGGATAGCTTCGGGAAGACAAACGTCAAACCTAACCAAAAATAA
Protein
MDVIVIGCGAAGVAALRKLHDAGLRVLGLEAADRIGGRICTIEYGDSTLDIGAAWCHGEKDNIVFELAEPLGLLGRPDPHDSWYVLSNGDLAPDATCKEILANIDEEVCKSHKNNVQSISQFVRNAVNTKETFKQFPRLTRSLLEVYERNNHLGGQDDPQHGKSLKGLDEHWPCEGEFLLNWRGRGYKTLLDVLLNKYPDPNEAIPVQILLNKHVECIRWGTSQPSHQISPLVQVKCTDGSLYAAKSVIVTLPLAVLKETHAQLFSPPLPQDKINSINSLHYCVLDKIYIEFTTPWWPKSAGKFVILWQEEDKAKFTKEEHWITEIYGLDPVQHQPNVLLAWIYGKGAEAMEKVSFDDLKAGIDKLLSIFKKKFPVTPVKSVLRSQWASNLLARSAYAYRCVATEENGASATTLSEPIYHGNGLPLVCFAGEATSYHRHSAVHGAVESGFREAQRLMDSFGKTNVKPNQK
Summary
Cofactor
FAD
Similarity
Belongs to the flavin monoamine oxidase family.
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family.
Uniprot
H9IVV4
A0A0N1PHN1
A0A194QDN3
A0A2A4JTY1
A0A437B2S8
A0A212ERF8
+ More
A0A2H1WCN3 A0A0L7L7C5 A0A0L7KUF4 A0A0L7KPT1 E2A8M5 U5EUZ7 A0A195D2Y1 A0A026X332 A0A1Y1NB58 Q16VW3 A0A023EU06 A0A1L8E4K2 A0A1I8Q9G4 A0A158NH52 A0A151JAS7 A0A1J1IYZ4 A0A0K8TNB2 A0A195F5U0 A0A3L8DLW3 A0A0C9R1M7 A0A151X5E5 A0A1Y9GK65 Q7QHJ2 A0A2A3ETS8 V9IIS9 A0A2J7RS87 A0A1W7R8F2 A0A088AF33 A0A1S4G756 A0A2M4ALI0 A0A1L8E4N5 A0A151J6B8 A0A2J7RSA6 A0A2M4BLW5 F4X074 A0A0L7QYF8 A0A2M4BLT9 Q2M0H7 A0A1S4FL64 A0A195D1J5 A0A3B0KFA7 A0A2A3EUE9 Q16WZ3 A0A1I8N8E6 A0A182MXI2 A0A182PI88 A0A182QD83 K7J8R0 A0A1B0C8L1 A0A182RCY9 A0A182X5H5 A0A182V8W6 A0A182LHM8 A0A453Z0A9 A0A2J7RS95 A0A158NHC7 A0A182TEZ7 A0A195AZ08 A0A0L0C2S0 B0WN37 A0A182WDD8 A0A084W3J4 A0A182LTM5 A0A182YEB5 A0A1B6G9D3 W5JHR8 A0A182S949 A0A182K9T3 A0A067R8V6 W8BV76 A0A182IQE9 F4X073 B4LEY2 A0A182FVI7 A0A1A9VI82 T1H025 A0A0P8XV49 A0A1S4FL53 A0A0R1DY17 A0A1B0AM68 A0A0J9RXG4 A0A1A9YKL1 U4UDG8 A0A0L0BM77 A0A1A9Z0N5 Q9VVK1 A0A1I8MY62 B0X0P7 A0A0Q5UJP0 A0A1A9WNE6 A0A1W4W496 Q16WZ5 B0WMR3
A0A2H1WCN3 A0A0L7L7C5 A0A0L7KUF4 A0A0L7KPT1 E2A8M5 U5EUZ7 A0A195D2Y1 A0A026X332 A0A1Y1NB58 Q16VW3 A0A023EU06 A0A1L8E4K2 A0A1I8Q9G4 A0A158NH52 A0A151JAS7 A0A1J1IYZ4 A0A0K8TNB2 A0A195F5U0 A0A3L8DLW3 A0A0C9R1M7 A0A151X5E5 A0A1Y9GK65 Q7QHJ2 A0A2A3ETS8 V9IIS9 A0A2J7RS87 A0A1W7R8F2 A0A088AF33 A0A1S4G756 A0A2M4ALI0 A0A1L8E4N5 A0A151J6B8 A0A2J7RSA6 A0A2M4BLW5 F4X074 A0A0L7QYF8 A0A2M4BLT9 Q2M0H7 A0A1S4FL64 A0A195D1J5 A0A3B0KFA7 A0A2A3EUE9 Q16WZ3 A0A1I8N8E6 A0A182MXI2 A0A182PI88 A0A182QD83 K7J8R0 A0A1B0C8L1 A0A182RCY9 A0A182X5H5 A0A182V8W6 A0A182LHM8 A0A453Z0A9 A0A2J7RS95 A0A158NHC7 A0A182TEZ7 A0A195AZ08 A0A0L0C2S0 B0WN37 A0A182WDD8 A0A084W3J4 A0A182LTM5 A0A182YEB5 A0A1B6G9D3 W5JHR8 A0A182S949 A0A182K9T3 A0A067R8V6 W8BV76 A0A182IQE9 F4X073 B4LEY2 A0A182FVI7 A0A1A9VI82 T1H025 A0A0P8XV49 A0A1S4FL53 A0A0R1DY17 A0A1B0AM68 A0A0J9RXG4 A0A1A9YKL1 U4UDG8 A0A0L0BM77 A0A1A9Z0N5 Q9VVK1 A0A1I8MY62 B0X0P7 A0A0Q5UJP0 A0A1A9WNE6 A0A1W4W496 Q16WZ5 B0WMR3
EC Number
1.4.3.-
Pubmed
19121390
26354079
22118469
26227816
20798317
24508170
+ More
28004739 17510324 24945155 21347285 26369729 30249741 12364791 21719571 15632085 17994087 25315136 20075255 20966253 26108605 24438588 25244985 20920257 23761445 24845553 24495485 17550304 22936249 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
28004739 17510324 24945155 21347285 26369729 30249741 12364791 21719571 15632085 17994087 25315136 20075255 20966253 26108605 24438588 25244985 20920257 23761445 24845553 24495485 17550304 22936249 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01006661
KQ459762
KPJ20122.1
KQ459193
KPJ03075.1
NWSH01000587
+ More
PCG75485.1 RSAL01000186 RVE44797.1 AGBW02013021 OWR44086.1 ODYU01007698 SOQ50716.1 JTDY01002453 KOB71383.1 JTDY01005742 KOB66676.1 JTDY01007622 KOB65071.1 GL437652 EFN70212.1 GANO01002099 JAB57772.1 KQ976973 KYN06719.1 KK107020 EZA62508.1 GEZM01010730 GEZM01010729 JAV93875.1 CH477579 EAT38750.1 GAPW01001212 JAC12386.1 GFDF01000406 JAV13678.1 ADTU01015583 ADTU01015584 ADTU01015585 KQ979264 KYN22062.1 CVRI01000064 CRL05312.1 GDAI01002178 JAI15425.1 KQ981768 KYN35838.1 QOIP01000006 RLU21371.1 GBYB01006752 JAG76519.1 KQ982510 KYQ55633.1 APCN01005438 AAAB01008816 EAA05268.4 KZ288185 PBC34904.1 JR049686 AEY61038.1 NEVH01000267 PNF43703.1 GEHC01000235 JAV47410.1 GGFK01008318 MBW41639.1 GFDF01000391 JAV13693.1 KQ979900 KYN18652.1 PNF43708.1 GGFJ01004904 MBW54045.1 GL888493 EGI60139.1 KQ414688 KOC63638.1 GGFJ01004905 MBW54046.1 CH379069 EAL30955.1 KYN06716.1 OUUW01000009 SPP84999.1 PBC34902.1 CH477549 EAT39128.1 AXCN02001709 AJWK01001026 AJWK01001027 AJWK01001028 PNF43706.1 ADTU01015577 ADTU01015578 ADTU01015579 KQ976698 KYM77478.1 JRES01001077 KNC25714.1 DS232006 EDS31456.1 ATLV01020055 KE525293 KFB44788.1 AXCM01003010 GECZ01010725 JAS59044.1 ADMH02001208 ETN63661.1 KK852624 KDR19928.1 GAMC01005687 GAMC01005686 JAC00869.1 EGI60138.1 CH940647 EDW69151.1 CAQQ02385011 CH902618 KPU78605.1 CM000159 KRK01990.1 JXJN01000364 CM002912 KMZ00298.1 KB632308 ERL92019.1 JRES01001669 KNC21028.1 AE014296 AAF49310.2 AGB94691.1 DS232242 EDS38289.1 CH954178 KQS44018.1 EAT39126.1 DS232000 EDS31148.1
PCG75485.1 RSAL01000186 RVE44797.1 AGBW02013021 OWR44086.1 ODYU01007698 SOQ50716.1 JTDY01002453 KOB71383.1 JTDY01005742 KOB66676.1 JTDY01007622 KOB65071.1 GL437652 EFN70212.1 GANO01002099 JAB57772.1 KQ976973 KYN06719.1 KK107020 EZA62508.1 GEZM01010730 GEZM01010729 JAV93875.1 CH477579 EAT38750.1 GAPW01001212 JAC12386.1 GFDF01000406 JAV13678.1 ADTU01015583 ADTU01015584 ADTU01015585 KQ979264 KYN22062.1 CVRI01000064 CRL05312.1 GDAI01002178 JAI15425.1 KQ981768 KYN35838.1 QOIP01000006 RLU21371.1 GBYB01006752 JAG76519.1 KQ982510 KYQ55633.1 APCN01005438 AAAB01008816 EAA05268.4 KZ288185 PBC34904.1 JR049686 AEY61038.1 NEVH01000267 PNF43703.1 GEHC01000235 JAV47410.1 GGFK01008318 MBW41639.1 GFDF01000391 JAV13693.1 KQ979900 KYN18652.1 PNF43708.1 GGFJ01004904 MBW54045.1 GL888493 EGI60139.1 KQ414688 KOC63638.1 GGFJ01004905 MBW54046.1 CH379069 EAL30955.1 KYN06716.1 OUUW01000009 SPP84999.1 PBC34902.1 CH477549 EAT39128.1 AXCN02001709 AJWK01001026 AJWK01001027 AJWK01001028 PNF43706.1 ADTU01015577 ADTU01015578 ADTU01015579 KQ976698 KYM77478.1 JRES01001077 KNC25714.1 DS232006 EDS31456.1 ATLV01020055 KE525293 KFB44788.1 AXCM01003010 GECZ01010725 JAS59044.1 ADMH02001208 ETN63661.1 KK852624 KDR19928.1 GAMC01005687 GAMC01005686 JAC00869.1 EGI60138.1 CH940647 EDW69151.1 CAQQ02385011 CH902618 KPU78605.1 CM000159 KRK01990.1 JXJN01000364 CM002912 KMZ00298.1 KB632308 ERL92019.1 JRES01001669 KNC21028.1 AE014296 AAF49310.2 AGB94691.1 DS232242 EDS38289.1 CH954178 KQS44018.1 EAT39126.1 DS232000 EDS31148.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000283053
UP000007151
+ More
UP000037510 UP000000311 UP000078542 UP000053097 UP000008820 UP000095300 UP000005205 UP000078492 UP000183832 UP000078541 UP000279307 UP000075809 UP000075840 UP000007062 UP000242457 UP000235965 UP000005203 UP000007755 UP000053825 UP000001819 UP000268350 UP000095301 UP000075884 UP000075885 UP000075886 UP000002358 UP000092461 UP000075900 UP000076407 UP000075903 UP000075882 UP000075902 UP000078540 UP000037069 UP000002320 UP000075920 UP000030765 UP000075883 UP000076408 UP000000673 UP000075901 UP000075881 UP000027135 UP000075880 UP000008792 UP000069272 UP000078200 UP000015102 UP000007801 UP000002282 UP000092460 UP000092443 UP000030742 UP000092445 UP000000803 UP000008711 UP000091820 UP000192221
UP000037510 UP000000311 UP000078542 UP000053097 UP000008820 UP000095300 UP000005205 UP000078492 UP000183832 UP000078541 UP000279307 UP000075809 UP000075840 UP000007062 UP000242457 UP000235965 UP000005203 UP000007755 UP000053825 UP000001819 UP000268350 UP000095301 UP000075884 UP000075885 UP000075886 UP000002358 UP000092461 UP000075900 UP000076407 UP000075903 UP000075882 UP000075902 UP000078540 UP000037069 UP000002320 UP000075920 UP000030765 UP000075883 UP000076408 UP000000673 UP000075901 UP000075881 UP000027135 UP000075880 UP000008792 UP000069272 UP000078200 UP000015102 UP000007801 UP000002282 UP000092460 UP000092443 UP000030742 UP000092445 UP000000803 UP000008711 UP000091820 UP000192221
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9IVV4
A0A0N1PHN1
A0A194QDN3
A0A2A4JTY1
A0A437B2S8
A0A212ERF8
+ More
A0A2H1WCN3 A0A0L7L7C5 A0A0L7KUF4 A0A0L7KPT1 E2A8M5 U5EUZ7 A0A195D2Y1 A0A026X332 A0A1Y1NB58 Q16VW3 A0A023EU06 A0A1L8E4K2 A0A1I8Q9G4 A0A158NH52 A0A151JAS7 A0A1J1IYZ4 A0A0K8TNB2 A0A195F5U0 A0A3L8DLW3 A0A0C9R1M7 A0A151X5E5 A0A1Y9GK65 Q7QHJ2 A0A2A3ETS8 V9IIS9 A0A2J7RS87 A0A1W7R8F2 A0A088AF33 A0A1S4G756 A0A2M4ALI0 A0A1L8E4N5 A0A151J6B8 A0A2J7RSA6 A0A2M4BLW5 F4X074 A0A0L7QYF8 A0A2M4BLT9 Q2M0H7 A0A1S4FL64 A0A195D1J5 A0A3B0KFA7 A0A2A3EUE9 Q16WZ3 A0A1I8N8E6 A0A182MXI2 A0A182PI88 A0A182QD83 K7J8R0 A0A1B0C8L1 A0A182RCY9 A0A182X5H5 A0A182V8W6 A0A182LHM8 A0A453Z0A9 A0A2J7RS95 A0A158NHC7 A0A182TEZ7 A0A195AZ08 A0A0L0C2S0 B0WN37 A0A182WDD8 A0A084W3J4 A0A182LTM5 A0A182YEB5 A0A1B6G9D3 W5JHR8 A0A182S949 A0A182K9T3 A0A067R8V6 W8BV76 A0A182IQE9 F4X073 B4LEY2 A0A182FVI7 A0A1A9VI82 T1H025 A0A0P8XV49 A0A1S4FL53 A0A0R1DY17 A0A1B0AM68 A0A0J9RXG4 A0A1A9YKL1 U4UDG8 A0A0L0BM77 A0A1A9Z0N5 Q9VVK1 A0A1I8MY62 B0X0P7 A0A0Q5UJP0 A0A1A9WNE6 A0A1W4W496 Q16WZ5 B0WMR3
A0A2H1WCN3 A0A0L7L7C5 A0A0L7KUF4 A0A0L7KPT1 E2A8M5 U5EUZ7 A0A195D2Y1 A0A026X332 A0A1Y1NB58 Q16VW3 A0A023EU06 A0A1L8E4K2 A0A1I8Q9G4 A0A158NH52 A0A151JAS7 A0A1J1IYZ4 A0A0K8TNB2 A0A195F5U0 A0A3L8DLW3 A0A0C9R1M7 A0A151X5E5 A0A1Y9GK65 Q7QHJ2 A0A2A3ETS8 V9IIS9 A0A2J7RS87 A0A1W7R8F2 A0A088AF33 A0A1S4G756 A0A2M4ALI0 A0A1L8E4N5 A0A151J6B8 A0A2J7RSA6 A0A2M4BLW5 F4X074 A0A0L7QYF8 A0A2M4BLT9 Q2M0H7 A0A1S4FL64 A0A195D1J5 A0A3B0KFA7 A0A2A3EUE9 Q16WZ3 A0A1I8N8E6 A0A182MXI2 A0A182PI88 A0A182QD83 K7J8R0 A0A1B0C8L1 A0A182RCY9 A0A182X5H5 A0A182V8W6 A0A182LHM8 A0A453Z0A9 A0A2J7RS95 A0A158NHC7 A0A182TEZ7 A0A195AZ08 A0A0L0C2S0 B0WN37 A0A182WDD8 A0A084W3J4 A0A182LTM5 A0A182YEB5 A0A1B6G9D3 W5JHR8 A0A182S949 A0A182K9T3 A0A067R8V6 W8BV76 A0A182IQE9 F4X073 B4LEY2 A0A182FVI7 A0A1A9VI82 T1H025 A0A0P8XV49 A0A1S4FL53 A0A0R1DY17 A0A1B0AM68 A0A0J9RXG4 A0A1A9YKL1 U4UDG8 A0A0L0BM77 A0A1A9Z0N5 Q9VVK1 A0A1I8MY62 B0X0P7 A0A0Q5UJP0 A0A1A9WNE6 A0A1W4W496 Q16WZ5 B0WMR3
PDB
5MBX
E-value=3.82743e-28,
Score=311
Ontologies
GO
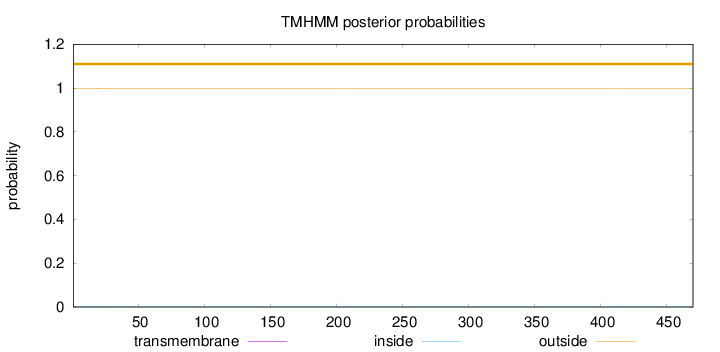
Topology
Length:
470
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01389
Exp number, first 60 AAs:
0.01255
Total prob of N-in:
0.00311
outside
1 - 470
Population Genetic Test Statistics
Pi
33.120705
Theta
28.162422
Tajima's D
0.5908
CLR
1.104348
CSRT
0.547772611369432
Interpretation
Uncertain
