Gene
KWMTBOMO12708
Annotation
hypothetical_protein_[Cotesia_congregata]
Full name
ATP-dependent DNA helicase
Location in the cell
Cytoplasmic Reliability : 1.735 Nuclear Reliability : 2.086
Sequence
CDS
ATGATGAAAGAGTTCGTTAAAGCATTGGATAAAAACAATGCTAGTTTTGAATACCTGTGTAAAAAAATTCCAAGATTATCTGATGCAAAAATTAAAGAAGGAGTGTTCGATGGACCGCAAATCAGATCTTTGATGGCTGATGAAAAATTTGATGTCACAATGAATAACACTGAATCAGATGCTTGGCTGGCGTTCAAAGATGCTGTCAACAATTTTCTAGGAAATAATAAACATCCCGACTATAAAAATAAGGTTGCAAACTTATTGGATAAGTATCAAAAATTGGGGTGTAATATGAGCATAAAACTCCACTTTCTAGATTCACATGTGGACTTTTTTCCTGATAATCTTGGTGATTACAGTGAAGAACAGGGAGAAAGGTTTCATCAGGACATAAAAACTATGGAGACAAGGTACCAAGGTCAATGGAATGTGAATATGATGGCTGATTACTGCTGGTCACTGACACGTGATATTACTGAAGATACTCACAAAAAAAACTACACCCAGACGTTATTTTGTCGCGAAACGTAA
Protein
MMKEFVKALDKNNASFEYLCKKIPRLSDAKIKEGVFDGPQIRSLMADEKFDVTMNNTESDAWLAFKDAVNNFLGNNKHPDYKNKVANLLDKYQKLGCNMSIKLHFLDSHVDFFPDNLGDYSEEQGERFHQDIKTMETRYQGQWNVNMMADYCWSLTRDITEDTHKKNYTQTLFCRET
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Uniprot
B9W4L2
A0A0V1H537
J9M0P4
A0A0J7K6Q9
A0A0J7K1T4
K7K0J2
+ More
D6WGQ1 A0A151I8N8 T1FLQ6 T1ET34 A0A151IZG3 A0A151JPZ6 A0A0V1MXV0 A0A151I634 A0A2J7QXK6 A0A2J7QG64 A0A0V1MYT4 A0A2J7PKD9 A0A433SPG2 A0A2J7R554 A0A0V1N5I0 V5GT02 A0A0J7KNJ2 T1FBV4 A0A2J7PUD9 A0A0J7KKE2 A0A2J7RBV7 A0A151IXS6 A0A2J7PET1 T1EMM0 T1F8E5 T1FC03 A0A151K2C4 A0A151J4T4 A0A151I7I3 A0A1B6K7B1 A0A3L8D898 T1EU09 A0A2S2PYM8 T1F7K8 T1ENK0 K7K089 T1F7P3 K7JGB9 A0A1B0CSA1 T1HHC3 T1H8T0 A0A1B6BZC3 T1HR57 T1I8M0 A0A151JQB4 T1EWI2 T1F4M2 X1WJT1 T1HKC5 R9XJ78 J9L6E9 A0A151JMX2 T1FLZ1 T1FK50 A0A3R7SWZ5 T1FFM5 T1F0J6 T1FC71 A0A0V1M1M0 T1F7E9 T1FJ14 T1F5V6 K7JVK8 K7JWZ1 K7JLC9 A0A0V1GPN4 B3SDN8 T1FI26 T1EWE5 A0A3L8D5E7
D6WGQ1 A0A151I8N8 T1FLQ6 T1ET34 A0A151IZG3 A0A151JPZ6 A0A0V1MXV0 A0A151I634 A0A2J7QXK6 A0A2J7QG64 A0A0V1MYT4 A0A2J7PKD9 A0A433SPG2 A0A2J7R554 A0A0V1N5I0 V5GT02 A0A0J7KNJ2 T1FBV4 A0A2J7PUD9 A0A0J7KKE2 A0A2J7RBV7 A0A151IXS6 A0A2J7PET1 T1EMM0 T1F8E5 T1FC03 A0A151K2C4 A0A151J4T4 A0A151I7I3 A0A1B6K7B1 A0A3L8D898 T1EU09 A0A2S2PYM8 T1F7K8 T1ENK0 K7K089 T1F7P3 K7JGB9 A0A1B0CSA1 T1HHC3 T1H8T0 A0A1B6BZC3 T1HR57 T1I8M0 A0A151JQB4 T1EWI2 T1F4M2 X1WJT1 T1HKC5 R9XJ78 J9L6E9 A0A151JMX2 T1FLZ1 T1FK50 A0A3R7SWZ5 T1FFM5 T1F0J6 T1FC71 A0A0V1M1M0 T1F7E9 T1FJ14 T1F5V6 K7JVK8 K7JWZ1 K7JLC9 A0A0V1GPN4 B3SDN8 T1FI26 T1EWE5 A0A3L8D5E7
EC Number
3.6.4.12
EMBL
FM212912
CAR82249.1
JYDP01000138
KRZ05510.1
ABLF02022650
LBMM01012852
+ More
KMQ85949.1 LBMM01016961 KMQ84254.1 AAZX01026046 KQ971327 EEZ99611.2 KQ978353 KYM94643.1 AMQM01011341 AMQM01011342 KB096270 ESO07053.1 AMQM01001161 KB097143 ESN99240.1 KQ980702 KYN14472.1 KQ978697 KYN29160.1 JYDO01000069 JYDO01000036 JYDO01000028 JYDO01000019 JYDO01000003 KRZ73025.1 KRZ75659.1 KRZ76474.1 KRZ77420.1 KRZ80366.1 KQ978517 KYM93378.1 NEVH01009379 PNF33316.1 NEVH01014836 PNF27561.1 JYDO01000024 KRZ76929.1 NEVH01024569 PNF16801.1 RQTK01001283 RUS71032.1 NEVH01007395 PNF35968.1 JYDO01000007 KRZ79249.1 GALX01003704 JAB64762.1 LBMM01005109 KMQ91809.1 AMQM01006096 KB097269 ESN97900.1 NEVH01021199 NEVH01019966 PNF19946.1 PNF22109.1 LBMM01006142 KMQ90883.1 NEVH01021935 NEVH01021574 NEVH01016945 NEVH01012564 NEVH01009765 NEVH01006566 NEVH01005890 NEVH01005312 NEVH01003746 NEVH01002687 NEVH01002549 NEVH01001362 PNF19214.1 PNF19612.1 PNF25043.1 PNF30218.1 PNF32869.1 PNF37912.1 PNF38301.1 PNF38709.1 PNF40208.1 PNF41665.1 PNF42166.1 PNF42683.1 KQ980792 KYN13106.1 NEVH01026088 NEVH01025127 NEVH01009083 NEVH01007402 NEVH01006727 NEVH01002562 PNF14850.1 PNF16142.1 PNF33735.1 PNF35867.1 PNF36872.1 PNF41992.1 AMQM01000003 KB095811 ESO11939.1 AMQM01005008 AMQM01005009 KB096743 ESO01734.1 ESO01735.1 AMQM01006116 ESN97976.1 LKEX01017198 KYN50270.1 KQ980100 KYN17766.1 KQ978418 KYM94050.1 GECU01000369 JAT07338.1 QOIP01000011 RLU16501.1 AMQM01001384 KB097495 ESN96365.1 GGMS01001300 MBY70503.1 AMQM01004807 KB096676 ESO03183.1 AMQM01000184 ESO12521.1 AAZX01005084 AMQM01004835 KB096716 ESO02739.1 AAZX01001882 AJWK01025825 ACPB03005055 ACPB03012626 GEDC01030686 GEDC01029533 GEDC01011220 JAS06612.1 JAS07765.1 JAS26078.1 ACPB03009529 ACPB03020207 KQ978650 KYN29423.1 AMQM01001999 KB097639 ESN93095.1 AMQM01003929 KB096365 ESO05370.1 ABLF02015932 ABLF02015935 ACPB03017840 EF710639 AGO14414.1 ABLF02005035 ABLF02005036 KQ978884 KYN27679.1 AMQM01012059 KB097190 ESN98204.1 AMQM01008975 KB095891 ESO10333.1 QCYY01001270 ROT79239.1 AMQM01007132 AMQM01007133 KB097572 ESN94079.1 AMQM01002958 KB095959 ESO09414.1 AMQM01006171 KB097304 ESN97698.1 JYDO01000323 KRZ65603.1 AMQM01004766 AMQM01004767 ESO03076.1 AMQM01008506 KB097788 ESN89970.1 AMQM01004332 KB096502 ESO04366.1 AAZX01003560 AAZX01025635 AAZX01026508 JYDP01000594 JYDP01000144 KRZ00058.1 KRZ05185.1 DS985282 EDV19166.1 AMQM01008140 KB097727 ESN91075.1 AMQM01001980 ESN93029.1 QOIP01000012 RLU15717.1
KMQ85949.1 LBMM01016961 KMQ84254.1 AAZX01026046 KQ971327 EEZ99611.2 KQ978353 KYM94643.1 AMQM01011341 AMQM01011342 KB096270 ESO07053.1 AMQM01001161 KB097143 ESN99240.1 KQ980702 KYN14472.1 KQ978697 KYN29160.1 JYDO01000069 JYDO01000036 JYDO01000028 JYDO01000019 JYDO01000003 KRZ73025.1 KRZ75659.1 KRZ76474.1 KRZ77420.1 KRZ80366.1 KQ978517 KYM93378.1 NEVH01009379 PNF33316.1 NEVH01014836 PNF27561.1 JYDO01000024 KRZ76929.1 NEVH01024569 PNF16801.1 RQTK01001283 RUS71032.1 NEVH01007395 PNF35968.1 JYDO01000007 KRZ79249.1 GALX01003704 JAB64762.1 LBMM01005109 KMQ91809.1 AMQM01006096 KB097269 ESN97900.1 NEVH01021199 NEVH01019966 PNF19946.1 PNF22109.1 LBMM01006142 KMQ90883.1 NEVH01021935 NEVH01021574 NEVH01016945 NEVH01012564 NEVH01009765 NEVH01006566 NEVH01005890 NEVH01005312 NEVH01003746 NEVH01002687 NEVH01002549 NEVH01001362 PNF19214.1 PNF19612.1 PNF25043.1 PNF30218.1 PNF32869.1 PNF37912.1 PNF38301.1 PNF38709.1 PNF40208.1 PNF41665.1 PNF42166.1 PNF42683.1 KQ980792 KYN13106.1 NEVH01026088 NEVH01025127 NEVH01009083 NEVH01007402 NEVH01006727 NEVH01002562 PNF14850.1 PNF16142.1 PNF33735.1 PNF35867.1 PNF36872.1 PNF41992.1 AMQM01000003 KB095811 ESO11939.1 AMQM01005008 AMQM01005009 KB096743 ESO01734.1 ESO01735.1 AMQM01006116 ESN97976.1 LKEX01017198 KYN50270.1 KQ980100 KYN17766.1 KQ978418 KYM94050.1 GECU01000369 JAT07338.1 QOIP01000011 RLU16501.1 AMQM01001384 KB097495 ESN96365.1 GGMS01001300 MBY70503.1 AMQM01004807 KB096676 ESO03183.1 AMQM01000184 ESO12521.1 AAZX01005084 AMQM01004835 KB096716 ESO02739.1 AAZX01001882 AJWK01025825 ACPB03005055 ACPB03012626 GEDC01030686 GEDC01029533 GEDC01011220 JAS06612.1 JAS07765.1 JAS26078.1 ACPB03009529 ACPB03020207 KQ978650 KYN29423.1 AMQM01001999 KB097639 ESN93095.1 AMQM01003929 KB096365 ESO05370.1 ABLF02015932 ABLF02015935 ACPB03017840 EF710639 AGO14414.1 ABLF02005035 ABLF02005036 KQ978884 KYN27679.1 AMQM01012059 KB097190 ESN98204.1 AMQM01008975 KB095891 ESO10333.1 QCYY01001270 ROT79239.1 AMQM01007132 AMQM01007133 KB097572 ESN94079.1 AMQM01002958 KB095959 ESO09414.1 AMQM01006171 KB097304 ESN97698.1 JYDO01000323 KRZ65603.1 AMQM01004766 AMQM01004767 ESO03076.1 AMQM01008506 KB097788 ESN89970.1 AMQM01004332 KB096502 ESO04366.1 AAZX01003560 AAZX01025635 AAZX01026508 JYDP01000594 JYDP01000144 KRZ00058.1 KRZ05185.1 DS985282 EDV19166.1 AMQM01008140 KB097727 ESN91075.1 AMQM01001980 ESN93029.1 QOIP01000012 RLU15717.1
Proteomes
ProteinModelPortal
B9W4L2
A0A0V1H537
J9M0P4
A0A0J7K6Q9
A0A0J7K1T4
K7K0J2
+ More
D6WGQ1 A0A151I8N8 T1FLQ6 T1ET34 A0A151IZG3 A0A151JPZ6 A0A0V1MXV0 A0A151I634 A0A2J7QXK6 A0A2J7QG64 A0A0V1MYT4 A0A2J7PKD9 A0A433SPG2 A0A2J7R554 A0A0V1N5I0 V5GT02 A0A0J7KNJ2 T1FBV4 A0A2J7PUD9 A0A0J7KKE2 A0A2J7RBV7 A0A151IXS6 A0A2J7PET1 T1EMM0 T1F8E5 T1FC03 A0A151K2C4 A0A151J4T4 A0A151I7I3 A0A1B6K7B1 A0A3L8D898 T1EU09 A0A2S2PYM8 T1F7K8 T1ENK0 K7K089 T1F7P3 K7JGB9 A0A1B0CSA1 T1HHC3 T1H8T0 A0A1B6BZC3 T1HR57 T1I8M0 A0A151JQB4 T1EWI2 T1F4M2 X1WJT1 T1HKC5 R9XJ78 J9L6E9 A0A151JMX2 T1FLZ1 T1FK50 A0A3R7SWZ5 T1FFM5 T1F0J6 T1FC71 A0A0V1M1M0 T1F7E9 T1FJ14 T1F5V6 K7JVK8 K7JWZ1 K7JLC9 A0A0V1GPN4 B3SDN8 T1FI26 T1EWE5 A0A3L8D5E7
D6WGQ1 A0A151I8N8 T1FLQ6 T1ET34 A0A151IZG3 A0A151JPZ6 A0A0V1MXV0 A0A151I634 A0A2J7QXK6 A0A2J7QG64 A0A0V1MYT4 A0A2J7PKD9 A0A433SPG2 A0A2J7R554 A0A0V1N5I0 V5GT02 A0A0J7KNJ2 T1FBV4 A0A2J7PUD9 A0A0J7KKE2 A0A2J7RBV7 A0A151IXS6 A0A2J7PET1 T1EMM0 T1F8E5 T1FC03 A0A151K2C4 A0A151J4T4 A0A151I7I3 A0A1B6K7B1 A0A3L8D898 T1EU09 A0A2S2PYM8 T1F7K8 T1ENK0 K7K089 T1F7P3 K7JGB9 A0A1B0CSA1 T1HHC3 T1H8T0 A0A1B6BZC3 T1HR57 T1I8M0 A0A151JQB4 T1EWI2 T1F4M2 X1WJT1 T1HKC5 R9XJ78 J9L6E9 A0A151JMX2 T1FLZ1 T1FK50 A0A3R7SWZ5 T1FFM5 T1F0J6 T1FC71 A0A0V1M1M0 T1F7E9 T1FJ14 T1F5V6 K7JVK8 K7JWZ1 K7JLC9 A0A0V1GPN4 B3SDN8 T1FI26 T1EWE5 A0A3L8D5E7
Ontologies
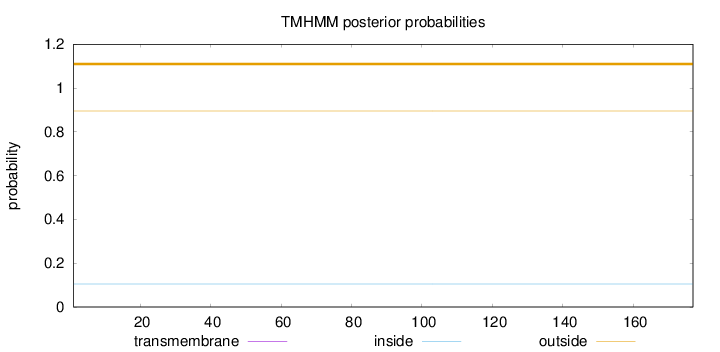
Topology
Length:
177
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10538
outside
1 - 177
Population Genetic Test Statistics
Pi
7.463944
Theta
7.77739
Tajima's D
-0.116066
CLR
0.370907
CSRT
0.346682665866707
Interpretation
Uncertain
