Gene
KWMTBOMO12704
Pre Gene Modal
BGIBMGA001600
Annotation
PREDICTED:_organic_cation_transporter_protein-like_isoform_X1_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.954
Sequence
CDS
ATGGAGGAAGGCAAGAAAGAAATCAATTTAGATTCGATACTGGAGAGCTTGGGTCCGTACGGGCGCTACAATTTACTGAATTTCGTGCTCCTACTGTACCCGGTCCTGCTGAGCAGCTTCTTCGAGTGCGGCTTTATATTCGAGGCTCAGGAACAGAATTACAGATGCGAAGTAGACGGTTGCGAGTTAAGGTTTAATGATACATCCTGGCTGCAGTACGCTATCCCGCTGGACGTGGAGAAACAGCGTCCTCAATCCTGCTTCATGTACCAGCAAGCAAACGTTACGAGTCAGCAGTGTTTGCCTGAAGACTTCATCAATAAAACTGTGCCTTGCGAGAGATTTGTTTACGAAAACGATTATTCTTTTGTCAAAGAGTTCAACCTTGTCTGCCAAGAATGGAAACGAGCGCTAGTCGGAACCATCCATAATTCTGGAATGTTTGTGGCCATGCTGTTCACTGGAGCCATATCGGATAGGTTCGGGAGGAAAGTTGCTGTAGGGTTTTCTTCCTGCGCAAGTTTCATATTCGGATTCGCTCGATCTTTCTCGCCGAGCTATCTGTTTTTCGTTGCCATGCATTTCCTGGAGGCTGGTTTCGGCGGAGGACTCTACCCTTCAGCTTATGTTTTCATTGTGGAACTGGTTGGCCTCAAGCAACGGGTCATCGTGTCGGCCATGTGCAACATGACCTTCGTTATCGGCGTCGTTCTCATAGCCCTTCTGGCCTGGGCTGTAAACAACTGGAGGACATACATCCTTATCCTCTACTGCCCCGCCACAATCGTCTTCGTATACGTTTGGTTTATGAACGAAAGCGCGCGCTGGCTTCTCAGCAAAGGACGAAGGGACGAAGCAATTGACGTACTGAAACGCGCCGCCAAGATCAACAATCTAGACCCGAGGAGTCTGGATCTAGATGCGCTCGGAGATCCCCTGCTGAAACCGCAGAACCAAACCGGAGACAAAAAGTCTCAGATGTACAAGGCCATCAGATCTGGTATAATATGGAAACGGGTCGCGATTTGCTCGTTTCTATGGATGACGTGTTCCCTAGTGTACTATGGTTTTTCCATTAATTCGGTTTCATTAAGTAGCAACAGATACGTTAGCTTCATGCTCGTGACGTTAGTGGAGATACCTGCCTATATTGTGGTAGTCATCGTGCTGGATAAATACGGGAGGAAGAAAACTTTATTGACGACTTATCTAACTTGTACGGTCATGAGTTTGCTGTTCGCTTTCTTGCCAAGATCTGATTACTGGTCGATACCCCTGTATCTAGTTGGGAAGTGGAGCGTCACATTAGCCTACAGTTCCGTCTATATCTACGTGTCGGAAGTCTTCCCTACTAACATGAGGCAGACTCTGATAAGCGTCTGCTCTACAGCTGGAAGGATCGGCTCTCTAATAGCTCCTTTAACGCCACTACTGTCATATTACCACAAATCGATGCCAACTGTGATCTTCGGAGCGATGTGCTTCATATCGAGCACTCTGGTCATGATGCTGCCGGAGACCAGGAACATGAGGCTTCCGGACACGATCGAGGAGGCCGAACACCTCTCCAGAATGAAAAGCAAGAACGGTGATACAACGGACTGA
Protein
MEEGKKEINLDSILESLGPYGRYNLLNFVLLLYPVLLSSFFECGFIFEAQEQNYRCEVDGCELRFNDTSWLQYAIPLDVEKQRPQSCFMYQQANVTSQQCLPEDFINKTVPCERFVYENDYSFVKEFNLVCQEWKRALVGTIHNSGMFVAMLFTGAISDRFGRKVAVGFSSCASFIFGFARSFSPSYLFFVAMHFLEAGFGGGLYPSAYVFIVELVGLKQRVIVSAMCNMTFVIGVVLIALLAWAVNNWRTYILILYCPATIVFVYVWFMNESARWLLSKGRRDEAIDVLKRAAKINNLDPRSLDLDALGDPLLKPQNQTGDKKSQMYKAIRSGIIWKRVAICSFLWMTCSLVYYGFSINSVSLSSNRYVSFMLVTLVEIPAYIVVVIVLDKYGRKKTLLTTYLTCTVMSLLFAFLPRSDYWSIPLYLVGKWSVTLAYSSVYIYVSEVFPTNMRQTLISVCSTAGRIGSLIAPLTPLLSYYHKSMPTVIFGAMCFISSTLVMMLPETRNMRLPDTIEEAEHLSRMKSKNGDTTD
Summary
Uniprot
A0A2H1VFS1
A0A2A4JC88
A0A212F2Q3
A0A194QC91
A0A3S2L3J0
A0A194RIS2
+ More
A0A2A4JBT3 A0A0L7LQ39 A0A0L7LKI8 A0A194QDP3 A0A212FPC5 A0A3S2NDN7 A0A2H1V1L3 A0A194REW9 A0A2A4J5Q3 A0A437B2S9 A0A0N1PI45 A0A194QI00 H9IWG7 A0A2A4J455 A0A212ERF4 A0A194QC06 A0A2H1V9B3 A0A2H1W2A4 A0A2H1VDM1 A0A2A4JU15 A0A3S2NA23 A0A212EJ20 A0A0L7KUG4 A0A2A4K5P1 A0A3S2NIQ2 A0A3S2M6B8 A0A2H1V9G0 Q16WK7 A0A194PSH8 A0A182GYP2 A0A182SYI8 A0A2A4K7E3 A0A067RSY2 A0A2A4K6M9 B0XDF0 A0A2A4K5R4 A0A182LZQ5 A0A194PT22 A0A2H1VA76 B0XDF1 A0A023ETG7 A0A182Y1J3 A0A084VS25 A0A182FXJ6 A0A2W1BRJ1 H9JI54 A0A182KSD5 U5EVF0 B0XDE9 A0A182QUL1 A0A2J7RPZ7 A0A194RGD2 A0A2J7RPY5 Q7Q0C3 A0A1Y1KD84 D2CG57 A0A2J7RPY7 A0A2H1V9D8 A0A2J7RPY0 A0A182Q0W8 A0A0V0GBF3 A0A212EJ24 A0A3S2TCP3 A0A182TM67 J3JWW6 A0A182Y1J2 A0A194RF43 H9IVN2 A0A1B6D578 A0A2W1BRL6 A0A194PJI6 A0A026W5E8 A0A3L8D7U5 A0A182KSC8 A0A2C9GR79 A0A2A4K5Q3 U5EWC3 A0A154PCE9 A0A182VDY0 A0A2H1VQB3 A0A182RFF8 A0A2A4J6J0 A0A182WEZ8 A0A084WCB2 A0A2M3ZIE3 A0A182WZR9 A0A0L7RIG1 A0A182M3B6 A0A2W1BMH3 A0A182FXJ5 H9JI53 A0A3S2NQS4 A0A0L7KV39
A0A2A4JBT3 A0A0L7LQ39 A0A0L7LKI8 A0A194QDP3 A0A212FPC5 A0A3S2NDN7 A0A2H1V1L3 A0A194REW9 A0A2A4J5Q3 A0A437B2S9 A0A0N1PI45 A0A194QI00 H9IWG7 A0A2A4J455 A0A212ERF4 A0A194QC06 A0A2H1V9B3 A0A2H1W2A4 A0A2H1VDM1 A0A2A4JU15 A0A3S2NA23 A0A212EJ20 A0A0L7KUG4 A0A2A4K5P1 A0A3S2NIQ2 A0A3S2M6B8 A0A2H1V9G0 Q16WK7 A0A194PSH8 A0A182GYP2 A0A182SYI8 A0A2A4K7E3 A0A067RSY2 A0A2A4K6M9 B0XDF0 A0A2A4K5R4 A0A182LZQ5 A0A194PT22 A0A2H1VA76 B0XDF1 A0A023ETG7 A0A182Y1J3 A0A084VS25 A0A182FXJ6 A0A2W1BRJ1 H9JI54 A0A182KSD5 U5EVF0 B0XDE9 A0A182QUL1 A0A2J7RPZ7 A0A194RGD2 A0A2J7RPY5 Q7Q0C3 A0A1Y1KD84 D2CG57 A0A2J7RPY7 A0A2H1V9D8 A0A2J7RPY0 A0A182Q0W8 A0A0V0GBF3 A0A212EJ24 A0A3S2TCP3 A0A182TM67 J3JWW6 A0A182Y1J2 A0A194RF43 H9IVN2 A0A1B6D578 A0A2W1BRL6 A0A194PJI6 A0A026W5E8 A0A3L8D7U5 A0A182KSC8 A0A2C9GR79 A0A2A4K5Q3 U5EWC3 A0A154PCE9 A0A182VDY0 A0A2H1VQB3 A0A182RFF8 A0A2A4J6J0 A0A182WEZ8 A0A084WCB2 A0A2M3ZIE3 A0A182WZR9 A0A0L7RIG1 A0A182M3B6 A0A2W1BMH3 A0A182FXJ5 H9JI53 A0A3S2NQS4 A0A0L7KV39
Pubmed
EMBL
ODYU01002328
SOQ39679.1
NWSH01002161
PCG69013.1
AGBW02010692
OWR48028.1
+ More
KQ459193 KPJ03082.1 RSAL01000186 RVE44802.1 KQ460325 KPJ15841.1 PCG69014.1 JTDY01000357 KOB77560.1 JTDY01000842 KOB75696.1 KPJ03085.1 AGBW02003000 OWR55596.1 RVE44804.1 ODYU01000079 SOQ34232.1 KPJ15840.1 NWSH01003151 PCG66854.1 RVE44801.1 KQ459762 KPJ20130.1 KPJ03081.1 BABH01006647 PCG66855.1 AGBW02013021 OWR44080.1 KPJ03083.1 ODYU01001366 SOQ37440.1 ODYU01005874 SOQ47209.1 ODYU01001802 SOQ38502.1 NWSH01000587 PCG75491.1 RSAL01000360 RVE42197.1 AGBW02014542 OWR41475.1 JTDY01005496 KOB66937.1 NWSH01000100 PCG79565.1 RSAL01000974 RVE41008.1 RSAL01000027 RVE51971.1 SOQ37441.1 CH477563 EAT38951.1 KQ459595 KPI95923.1 JXUM01021216 KQ560594 KXJ81736.1 PCG79570.1 KK852482 KDR22929.1 PCG79564.1 DS232756 EDS45428.1 PCG79571.1 AXCM01003135 KPI95914.1 ODYU01001220 SOQ37134.1 EDS45429.1 GAPW01001000 JAC12598.1 ATLV01015792 KE525036 KFB40769.1 KZ149964 PZC76264.1 BABH01041118 BABH01041119 GANO01001906 JAB57965.1 EDS45427.1 AXCN02001021 NEVH01001354 PNF42898.1 KQ460205 KPJ16878.1 PNF42894.1 AAAB01008986 EAA00192.3 GEZM01087818 JAV58458.1 KQ972004 EFA12973.2 PNF42889.1 SOQ37439.1 PNF42891.1 GECL01000840 JAP05284.1 OWR41479.1 RVE42200.1 APGK01035527 APGK01035528 BT127734 KB740927 KB631762 AEE62696.1 ENN78004.1 ERL85926.1 KQ460313 KPJ16069.1 BABH01039623 GEDC01016480 GEDC01008507 JAS20818.1 JAS28791.1 PZC76255.1 KQ459606 KPI91250.1 KK107403 EZA51300.1 QOIP01000011 RLU16525.1 APCN01003305 PCG79561.1 GANO01000632 JAB59239.1 KQ434870 KZC09576.1 ODYU01003788 SOQ42987.1 NWSH01002815 PCG67489.1 ATLV01022651 KE525335 KFB47856.1 GGFM01007521 MBW28272.1 KQ414585 KOC70516.1 PZC76259.1 BABH01041117 RVE42198.1 KOB66936.1
KQ459193 KPJ03082.1 RSAL01000186 RVE44802.1 KQ460325 KPJ15841.1 PCG69014.1 JTDY01000357 KOB77560.1 JTDY01000842 KOB75696.1 KPJ03085.1 AGBW02003000 OWR55596.1 RVE44804.1 ODYU01000079 SOQ34232.1 KPJ15840.1 NWSH01003151 PCG66854.1 RVE44801.1 KQ459762 KPJ20130.1 KPJ03081.1 BABH01006647 PCG66855.1 AGBW02013021 OWR44080.1 KPJ03083.1 ODYU01001366 SOQ37440.1 ODYU01005874 SOQ47209.1 ODYU01001802 SOQ38502.1 NWSH01000587 PCG75491.1 RSAL01000360 RVE42197.1 AGBW02014542 OWR41475.1 JTDY01005496 KOB66937.1 NWSH01000100 PCG79565.1 RSAL01000974 RVE41008.1 RSAL01000027 RVE51971.1 SOQ37441.1 CH477563 EAT38951.1 KQ459595 KPI95923.1 JXUM01021216 KQ560594 KXJ81736.1 PCG79570.1 KK852482 KDR22929.1 PCG79564.1 DS232756 EDS45428.1 PCG79571.1 AXCM01003135 KPI95914.1 ODYU01001220 SOQ37134.1 EDS45429.1 GAPW01001000 JAC12598.1 ATLV01015792 KE525036 KFB40769.1 KZ149964 PZC76264.1 BABH01041118 BABH01041119 GANO01001906 JAB57965.1 EDS45427.1 AXCN02001021 NEVH01001354 PNF42898.1 KQ460205 KPJ16878.1 PNF42894.1 AAAB01008986 EAA00192.3 GEZM01087818 JAV58458.1 KQ972004 EFA12973.2 PNF42889.1 SOQ37439.1 PNF42891.1 GECL01000840 JAP05284.1 OWR41479.1 RVE42200.1 APGK01035527 APGK01035528 BT127734 KB740927 KB631762 AEE62696.1 ENN78004.1 ERL85926.1 KQ460313 KPJ16069.1 BABH01039623 GEDC01016480 GEDC01008507 JAS20818.1 JAS28791.1 PZC76255.1 KQ459606 KPI91250.1 KK107403 EZA51300.1 QOIP01000011 RLU16525.1 APCN01003305 PCG79561.1 GANO01000632 JAB59239.1 KQ434870 KZC09576.1 ODYU01003788 SOQ42987.1 NWSH01002815 PCG67489.1 ATLV01022651 KE525335 KFB47856.1 GGFM01007521 MBW28272.1 KQ414585 KOC70516.1 PZC76259.1 BABH01041117 RVE42198.1 KOB66936.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000283053
UP000053240
UP000037510
+ More
UP000005204 UP000008820 UP000069940 UP000249989 UP000075901 UP000027135 UP000002320 UP000075883 UP000076408 UP000030765 UP000069272 UP000075882 UP000075886 UP000235965 UP000007062 UP000007266 UP000075902 UP000019118 UP000030742 UP000053097 UP000279307 UP000075840 UP000076502 UP000075903 UP000075900 UP000075920 UP000076407 UP000053825
UP000005204 UP000008820 UP000069940 UP000249989 UP000075901 UP000027135 UP000002320 UP000075883 UP000076408 UP000030765 UP000069272 UP000075882 UP000075886 UP000235965 UP000007062 UP000007266 UP000075902 UP000019118 UP000030742 UP000053097 UP000279307 UP000075840 UP000076502 UP000075903 UP000075900 UP000075920 UP000076407 UP000053825
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2H1VFS1
A0A2A4JC88
A0A212F2Q3
A0A194QC91
A0A3S2L3J0
A0A194RIS2
+ More
A0A2A4JBT3 A0A0L7LQ39 A0A0L7LKI8 A0A194QDP3 A0A212FPC5 A0A3S2NDN7 A0A2H1V1L3 A0A194REW9 A0A2A4J5Q3 A0A437B2S9 A0A0N1PI45 A0A194QI00 H9IWG7 A0A2A4J455 A0A212ERF4 A0A194QC06 A0A2H1V9B3 A0A2H1W2A4 A0A2H1VDM1 A0A2A4JU15 A0A3S2NA23 A0A212EJ20 A0A0L7KUG4 A0A2A4K5P1 A0A3S2NIQ2 A0A3S2M6B8 A0A2H1V9G0 Q16WK7 A0A194PSH8 A0A182GYP2 A0A182SYI8 A0A2A4K7E3 A0A067RSY2 A0A2A4K6M9 B0XDF0 A0A2A4K5R4 A0A182LZQ5 A0A194PT22 A0A2H1VA76 B0XDF1 A0A023ETG7 A0A182Y1J3 A0A084VS25 A0A182FXJ6 A0A2W1BRJ1 H9JI54 A0A182KSD5 U5EVF0 B0XDE9 A0A182QUL1 A0A2J7RPZ7 A0A194RGD2 A0A2J7RPY5 Q7Q0C3 A0A1Y1KD84 D2CG57 A0A2J7RPY7 A0A2H1V9D8 A0A2J7RPY0 A0A182Q0W8 A0A0V0GBF3 A0A212EJ24 A0A3S2TCP3 A0A182TM67 J3JWW6 A0A182Y1J2 A0A194RF43 H9IVN2 A0A1B6D578 A0A2W1BRL6 A0A194PJI6 A0A026W5E8 A0A3L8D7U5 A0A182KSC8 A0A2C9GR79 A0A2A4K5Q3 U5EWC3 A0A154PCE9 A0A182VDY0 A0A2H1VQB3 A0A182RFF8 A0A2A4J6J0 A0A182WEZ8 A0A084WCB2 A0A2M3ZIE3 A0A182WZR9 A0A0L7RIG1 A0A182M3B6 A0A2W1BMH3 A0A182FXJ5 H9JI53 A0A3S2NQS4 A0A0L7KV39
A0A2A4JBT3 A0A0L7LQ39 A0A0L7LKI8 A0A194QDP3 A0A212FPC5 A0A3S2NDN7 A0A2H1V1L3 A0A194REW9 A0A2A4J5Q3 A0A437B2S9 A0A0N1PI45 A0A194QI00 H9IWG7 A0A2A4J455 A0A212ERF4 A0A194QC06 A0A2H1V9B3 A0A2H1W2A4 A0A2H1VDM1 A0A2A4JU15 A0A3S2NA23 A0A212EJ20 A0A0L7KUG4 A0A2A4K5P1 A0A3S2NIQ2 A0A3S2M6B8 A0A2H1V9G0 Q16WK7 A0A194PSH8 A0A182GYP2 A0A182SYI8 A0A2A4K7E3 A0A067RSY2 A0A2A4K6M9 B0XDF0 A0A2A4K5R4 A0A182LZQ5 A0A194PT22 A0A2H1VA76 B0XDF1 A0A023ETG7 A0A182Y1J3 A0A084VS25 A0A182FXJ6 A0A2W1BRJ1 H9JI54 A0A182KSD5 U5EVF0 B0XDE9 A0A182QUL1 A0A2J7RPZ7 A0A194RGD2 A0A2J7RPY5 Q7Q0C3 A0A1Y1KD84 D2CG57 A0A2J7RPY7 A0A2H1V9D8 A0A2J7RPY0 A0A182Q0W8 A0A0V0GBF3 A0A212EJ24 A0A3S2TCP3 A0A182TM67 J3JWW6 A0A182Y1J2 A0A194RF43 H9IVN2 A0A1B6D578 A0A2W1BRL6 A0A194PJI6 A0A026W5E8 A0A3L8D7U5 A0A182KSC8 A0A2C9GR79 A0A2A4K5Q3 U5EWC3 A0A154PCE9 A0A182VDY0 A0A2H1VQB3 A0A182RFF8 A0A2A4J6J0 A0A182WEZ8 A0A084WCB2 A0A2M3ZIE3 A0A182WZR9 A0A0L7RIG1 A0A182M3B6 A0A2W1BMH3 A0A182FXJ5 H9JI53 A0A3S2NQS4 A0A0L7KV39
PDB
4LDS
E-value=6.01895e-08,
Score=138
Ontologies
GO
Topology
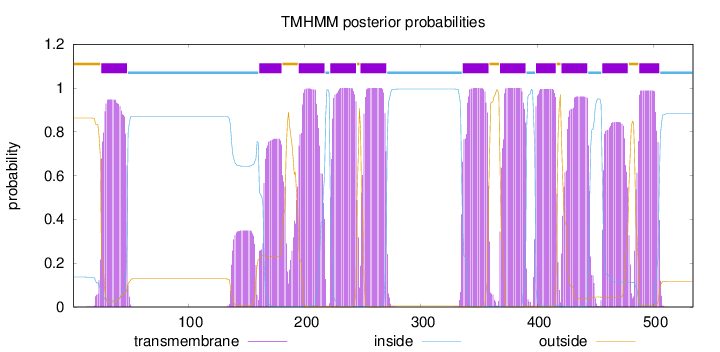
Length:
534
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
231.00292
Exp number, first 60 AAs:
20.96708
Total prob of N-in:
0.13656
POSSIBLE N-term signal
sequence
outside
1 - 24
TMhelix
25 - 47
inside
48 - 160
TMhelix
161 - 180
outside
181 - 194
TMhelix
195 - 217
inside
218 - 221
TMhelix
222 - 244
outside
245 - 247
TMhelix
248 - 270
inside
271 - 335
TMhelix
336 - 358
outside
359 - 367
TMhelix
368 - 390
inside
391 - 398
TMhelix
399 - 416
outside
417 - 420
TMhelix
421 - 443
inside
444 - 455
TMhelix
456 - 478
outside
479 - 487
TMhelix
488 - 505
inside
506 - 534
Population Genetic Test Statistics
Pi
3.496589
Theta
19.314283
Tajima's D
-1.432678
CLR
1.807464
CSRT
0.0690465476726164
Interpretation
Uncertain
