Gene
KWMTBOMO12703
Pre Gene Modal
BGIBMGA001599
Annotation
PREDICTED:_organic_cation_transporter_protein-like_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.882
Sequence
CDS
ATGACGGACACTAACGCCGGCGGACTGGACACCGTTTTAAAGCAACTAGGTCCTTTTGGGAGATATAACGTTATCAATTACATGTTATTCTTATATCCCTGTTTTTTGGCCGGAATGTATGGATCGGTTTATGTATTCGAAGCTTCCGATATTAATTATAGATGCGAAATAAAAGAATGCGAAACATCTAACAACAGCGTTTGGATCGAATTCGGGATACCACAGGACAAAGGTAGTTTGTCTAAATGCGAAAGGTACTCGTTCAAAAATAGCACATTGAATAATGGAACTTGTACTCAAGATATGTTCGACAATCATCTAAGAGAAAAATGTGATACTTATGTGTACAGCGACGAAGATTCGGCAGTTAAAGATTTCAATCTGGGTTGCGAAAATTGGAAGAGGACACTGATCGGTACAATCCACAACTCTGGTCTATTTGTGTCTCTACCTCTCACCGGTATCATGTCTGATAGATTCGGTAGGAAAAAGGCTTTGGCTGTAGCTTCACTGATGAACTGCATTTTTGGAGTGATGAGATCGTTTTCAGTCAACTACCTTATGATGGTAGCTTGCGAGTTCTGCGAGGCGGCTTTTGGCGCTGGGGCATATTCGACAGCTTTTGTTTTAGCAATGGAACTCGTAGGACCCAAAGGCAGAGTATTCGGGAATACTTTGATAAATGTATTCTACGTTTGTGGCTTAATGACATTGGCTGGTTTATCATGGGCACTACAAGACTGGAGAACCTTACTGCGAGTCATATACTCGCCAGCATGCCTTGTCTTCTCCTACATTTGGATTCTAAACGAGAGCATAAGATGGTTGCTCAGTAAGGGAAAACACGAAAAGGCCGTCGAAATACTCAGAAAGGCGGCAAAAATGAATAAAGTGGAACTATCTGAGCAAGATCTAACGCCAATTTATGAACTAGATAAAGTCAAAGCGACAAAAGATAAAGATTATCTCGTTGTAAGTGCCGAAGGTCCATCTATCTTCCAGCAAGTGATCAGATCGAGTATAATCATCAAGAGATTATTAACGTGCTCCTATCTTTGGATTACTTGCACTTTCGTGTACTACGGATTGTCAATAAACTCTGTCTCGTTGGCTGGTAATAAATACGTAAATTTTATGCTGGTTGCCTTCGTGGAGATTCCAGCGAACTTTGCGTGTCTGGCCGTTTTGGATCGATTCGGAAGGAAGAGGGTTTTGATAACGATGTACATTTTAAGCGCGATTCTGTGCATTGGATTGGCTTTTATCCCTAAAGATAAAGCATGGTGGTCCTTGATCGTATATCTCTCTGGAAAGTTCTCGATAACGATCGCGTACAGTTCCGTCTACATCTACGTGTCAGAAGTCTTCCCTACGAATGTGAGGCAGTCACTTCTAGCCGTCTGCTCGTCTACTGGACGTATTGGGTCAACTTTGGCACCTTTAACACCTCTGCTCGCTCTATACTACGAGAAACTCCCGGTCATTTTCTTCGGTACTTTAGCAGTTTTGGCCGGCATGCTGGTCTTCACGCTGCCGGAAACTATAAACGTTCGGTTACCAGACACGATAGAGGAGGCCGAGAGATTGTCTAATCGTAAAAACGAAGAAAACCTTTCGTGA
Protein
MTDTNAGGLDTVLKQLGPFGRYNVINYMLFLYPCFLAGMYGSVYVFEASDINYRCEIKECETSNNSVWIEFGIPQDKGSLSKCERYSFKNSTLNNGTCTQDMFDNHLREKCDTYVYSDEDSAVKDFNLGCENWKRTLIGTIHNSGLFVSLPLTGIMSDRFGRKKALAVASLMNCIFGVMRSFSVNYLMMVACEFCEAAFGAGAYSTAFVLAMELVGPKGRVFGNTLINVFYVCGLMTLAGLSWALQDWRTLLRVIYSPACLVFSYIWILNESIRWLLSKGKHEKAVEILRKAAKMNKVELSEQDLTPIYELDKVKATKDKDYLVVSAEGPSIFQQVIRSSIIIKRLLTCSYLWITCTFVYYGLSINSVSLAGNKYVNFMLVAFVEIPANFACLAVLDRFGRKRVLITMYILSAILCIGLAFIPKDKAWWSLIVYLSGKFSITIAYSSVYIYVSEVFPTNVRQSLLAVCSSTGRIGSTLAPLTPLLALYYEKLPVIFFGTLAVLAGMLVFTLPETINVRLPDTIEEAERLSNRKNEENLS
Summary
Uniprot
H9IWG7
A0A0L7LKI8
A0A194QDP3
A0A194REW9
A0A212FPC5
A0A3S2NDN7
+ More
A0A2A4J5Q3 A0A2H1V1L3 A0A2A4J455 A0A194QC06 A0A437B2S9 A0A0N1PI45 A0A3S2L3J0 A0A194QI00 A0A194QC91 A0A2A4JC88 A0A2H1VFS1 A0A194RIS2 A0A3S2NIQ2 A0A3S2M6B8 A0A212F2Q3 A0A212ERF4 A0A212EJ20 A0A1B6D578 A0A2H1V9B3 A0A067RSY2 A0A3S2NA23 A0A2J7RPY7 A0A2J7RPY0 A0A2W1BRJ1 A0A2A4K5R4 A0A2J7RPZ7 A0A2H1VA76 A0A194RF43 A0A2H1W2A4 A0A2H1VDM1 A0A2A4K7E3 A0A2A4JU15 A0A194PT22 A0A182GYP2 A0A0L7RIG1 A0A154PCE9 B0XDF1 Q16WK7 A0A088AAF9 V9ICM7 A0A195DF58 A0A2A3E3H4 A0A067RDE7 A0A2J7RPY5 A0A2A4K5Q2 A0A158NQD1 A0A1L8DEQ3 A0A195CVD4 A0A1L8DEI6 A0A0V0GBF3 D2CG57 A0A182LZQ5 A0A3L8D7U5 A0A194PSH8 U5EWC3 A0A3S2TCP3 A0A2J7RPY4 A0A2W1BMH3 B0XDE9 A0A194PR44 A0A151WZG1 A0A0L7KTQ3 A0A195E542 A0A182SYI8 U5EVF0 A0A182KSD5 A0A067RGL9 A0A026W5E8 A0A023ETG7 A0A212EJ24 A0A3S2L9U7 A0A151WY09 A0A437AVL7 B0XDF0 A0A1Y1KD84 A0A182Y1J3 H9IVN2 E2A7K4 A0A1I8MQZ5 A0A2H1V9D8 A0A084WCB2 Q7Q0C3 A0A212F848 A0A182QUL1 A0A195D353 A0A1B0DMI4 A0A2A4K5Q3 A0A182J645 A0A195ETW6 A0A2A4K6M9 Q2M044 A0A0A1WIH3 W8BAQ3 A0A2W1BRL6
A0A2A4J5Q3 A0A2H1V1L3 A0A2A4J455 A0A194QC06 A0A437B2S9 A0A0N1PI45 A0A3S2L3J0 A0A194QI00 A0A194QC91 A0A2A4JC88 A0A2H1VFS1 A0A194RIS2 A0A3S2NIQ2 A0A3S2M6B8 A0A212F2Q3 A0A212ERF4 A0A212EJ20 A0A1B6D578 A0A2H1V9B3 A0A067RSY2 A0A3S2NA23 A0A2J7RPY7 A0A2J7RPY0 A0A2W1BRJ1 A0A2A4K5R4 A0A2J7RPZ7 A0A2H1VA76 A0A194RF43 A0A2H1W2A4 A0A2H1VDM1 A0A2A4K7E3 A0A2A4JU15 A0A194PT22 A0A182GYP2 A0A0L7RIG1 A0A154PCE9 B0XDF1 Q16WK7 A0A088AAF9 V9ICM7 A0A195DF58 A0A2A3E3H4 A0A067RDE7 A0A2J7RPY5 A0A2A4K5Q2 A0A158NQD1 A0A1L8DEQ3 A0A195CVD4 A0A1L8DEI6 A0A0V0GBF3 D2CG57 A0A182LZQ5 A0A3L8D7U5 A0A194PSH8 U5EWC3 A0A3S2TCP3 A0A2J7RPY4 A0A2W1BMH3 B0XDE9 A0A194PR44 A0A151WZG1 A0A0L7KTQ3 A0A195E542 A0A182SYI8 U5EVF0 A0A182KSD5 A0A067RGL9 A0A026W5E8 A0A023ETG7 A0A212EJ24 A0A3S2L9U7 A0A151WY09 A0A437AVL7 B0XDF0 A0A1Y1KD84 A0A182Y1J3 H9IVN2 E2A7K4 A0A1I8MQZ5 A0A2H1V9D8 A0A084WCB2 Q7Q0C3 A0A212F848 A0A182QUL1 A0A195D353 A0A1B0DMI4 A0A2A4K5Q3 A0A182J645 A0A195ETW6 A0A2A4K6M9 Q2M044 A0A0A1WIH3 W8BAQ3 A0A2W1BRL6
Pubmed
EMBL
BABH01006647
JTDY01000842
KOB75696.1
KQ459193
KPJ03085.1
KQ460325
+ More
KPJ15840.1 AGBW02003000 OWR55596.1 RSAL01000186 RVE44804.1 NWSH01003151 PCG66854.1 ODYU01000079 SOQ34232.1 PCG66855.1 KPJ03083.1 RVE44801.1 KQ459762 KPJ20130.1 RVE44802.1 KPJ03081.1 KPJ03082.1 NWSH01002161 PCG69013.1 ODYU01002328 SOQ39679.1 KPJ15841.1 RSAL01000974 RVE41008.1 RSAL01000027 RVE51971.1 AGBW02010692 OWR48028.1 AGBW02013021 OWR44080.1 AGBW02014542 OWR41475.1 GEDC01016480 GEDC01008507 JAS20818.1 JAS28791.1 ODYU01001366 SOQ37440.1 KK852482 KDR22929.1 RSAL01000360 RVE42197.1 NEVH01001354 PNF42889.1 PNF42891.1 KZ149964 PZC76264.1 NWSH01000100 PCG79571.1 PNF42898.1 ODYU01001220 SOQ37134.1 KQ460313 KPJ16069.1 ODYU01005874 SOQ47209.1 ODYU01001802 SOQ38502.1 PCG79570.1 NWSH01000587 PCG75491.1 KQ459595 KPI95914.1 JXUM01021216 KQ560594 KXJ81736.1 KQ414585 KOC70516.1 KQ434870 KZC09576.1 DS232756 EDS45429.1 CH477563 EAT38951.1 JR038305 AEY58206.1 KQ980905 KYN11533.1 KZ288444 PBC25689.1 KK852530 KDR21906.1 PNF42894.1 PCG79557.1 ADTU01001734 GFDF01009227 JAV04857.1 KQ977259 KYN04595.1 GFDF01009228 JAV04856.1 GECL01000840 JAP05284.1 KQ972004 EFA12973.2 AXCM01003135 QOIP01000011 RLU16525.1 KPI95923.1 GANO01000632 JAB59239.1 RVE42200.1 PNF42897.1 PZC76259.1 EDS45427.1 KPI95921.1 KQ982639 KYQ53259.1 JTDY01005909 KOB66490.1 KQ979657 KYN19974.1 GANO01001906 JAB57965.1 KDR22927.1 KK107403 EZA51300.1 GAPW01001000 JAC12598.1 OWR41479.1 RVE42203.1 KQ982662 KYQ52637.1 RVE42199.1 EDS45428.1 GEZM01087818 JAV58458.1 BABH01039623 GL437348 EFN70576.1 SOQ37439.1 ATLV01022651 KE525335 KFB47856.1 AAAB01008986 EAA00192.3 AGBW02009797 OWR49901.1 AXCN02001021 KQ976885 KYN07327.1 AJVK01006968 PCG79561.1 KQ981979 KYN31324.1 PCG79564.1 CH379069 EAL31092.3 GBXI01016094 JAC98197.1 GAMC01016374 JAB90181.1 PZC76255.1
KPJ15840.1 AGBW02003000 OWR55596.1 RSAL01000186 RVE44804.1 NWSH01003151 PCG66854.1 ODYU01000079 SOQ34232.1 PCG66855.1 KPJ03083.1 RVE44801.1 KQ459762 KPJ20130.1 RVE44802.1 KPJ03081.1 KPJ03082.1 NWSH01002161 PCG69013.1 ODYU01002328 SOQ39679.1 KPJ15841.1 RSAL01000974 RVE41008.1 RSAL01000027 RVE51971.1 AGBW02010692 OWR48028.1 AGBW02013021 OWR44080.1 AGBW02014542 OWR41475.1 GEDC01016480 GEDC01008507 JAS20818.1 JAS28791.1 ODYU01001366 SOQ37440.1 KK852482 KDR22929.1 RSAL01000360 RVE42197.1 NEVH01001354 PNF42889.1 PNF42891.1 KZ149964 PZC76264.1 NWSH01000100 PCG79571.1 PNF42898.1 ODYU01001220 SOQ37134.1 KQ460313 KPJ16069.1 ODYU01005874 SOQ47209.1 ODYU01001802 SOQ38502.1 PCG79570.1 NWSH01000587 PCG75491.1 KQ459595 KPI95914.1 JXUM01021216 KQ560594 KXJ81736.1 KQ414585 KOC70516.1 KQ434870 KZC09576.1 DS232756 EDS45429.1 CH477563 EAT38951.1 JR038305 AEY58206.1 KQ980905 KYN11533.1 KZ288444 PBC25689.1 KK852530 KDR21906.1 PNF42894.1 PCG79557.1 ADTU01001734 GFDF01009227 JAV04857.1 KQ977259 KYN04595.1 GFDF01009228 JAV04856.1 GECL01000840 JAP05284.1 KQ972004 EFA12973.2 AXCM01003135 QOIP01000011 RLU16525.1 KPI95923.1 GANO01000632 JAB59239.1 RVE42200.1 PNF42897.1 PZC76259.1 EDS45427.1 KPI95921.1 KQ982639 KYQ53259.1 JTDY01005909 KOB66490.1 KQ979657 KYN19974.1 GANO01001906 JAB57965.1 KDR22927.1 KK107403 EZA51300.1 GAPW01001000 JAC12598.1 OWR41479.1 RVE42203.1 KQ982662 KYQ52637.1 RVE42199.1 EDS45428.1 GEZM01087818 JAV58458.1 BABH01039623 GL437348 EFN70576.1 SOQ37439.1 ATLV01022651 KE525335 KFB47856.1 AAAB01008986 EAA00192.3 AGBW02009797 OWR49901.1 AXCN02001021 KQ976885 KYN07327.1 AJVK01006968 PCG79561.1 KQ981979 KYN31324.1 PCG79564.1 CH379069 EAL31092.3 GBXI01016094 JAC98197.1 GAMC01016374 JAB90181.1 PZC76255.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000218220 UP000027135 UP000235965 UP000069940 UP000249989 UP000053825 UP000076502 UP000002320 UP000008820 UP000005203 UP000078492 UP000242457 UP000005205 UP000078542 UP000007266 UP000075883 UP000279307 UP000075809 UP000075901 UP000075882 UP000053097 UP000076408 UP000000311 UP000095301 UP000030765 UP000007062 UP000075886 UP000092462 UP000075880 UP000078541 UP000001819
UP000218220 UP000027135 UP000235965 UP000069940 UP000249989 UP000053825 UP000076502 UP000002320 UP000008820 UP000005203 UP000078492 UP000242457 UP000005205 UP000078542 UP000007266 UP000075883 UP000279307 UP000075809 UP000075901 UP000075882 UP000053097 UP000076408 UP000000311 UP000095301 UP000030765 UP000007062 UP000075886 UP000092462 UP000075880 UP000078541 UP000001819
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9IWG7
A0A0L7LKI8
A0A194QDP3
A0A194REW9
A0A212FPC5
A0A3S2NDN7
+ More
A0A2A4J5Q3 A0A2H1V1L3 A0A2A4J455 A0A194QC06 A0A437B2S9 A0A0N1PI45 A0A3S2L3J0 A0A194QI00 A0A194QC91 A0A2A4JC88 A0A2H1VFS1 A0A194RIS2 A0A3S2NIQ2 A0A3S2M6B8 A0A212F2Q3 A0A212ERF4 A0A212EJ20 A0A1B6D578 A0A2H1V9B3 A0A067RSY2 A0A3S2NA23 A0A2J7RPY7 A0A2J7RPY0 A0A2W1BRJ1 A0A2A4K5R4 A0A2J7RPZ7 A0A2H1VA76 A0A194RF43 A0A2H1W2A4 A0A2H1VDM1 A0A2A4K7E3 A0A2A4JU15 A0A194PT22 A0A182GYP2 A0A0L7RIG1 A0A154PCE9 B0XDF1 Q16WK7 A0A088AAF9 V9ICM7 A0A195DF58 A0A2A3E3H4 A0A067RDE7 A0A2J7RPY5 A0A2A4K5Q2 A0A158NQD1 A0A1L8DEQ3 A0A195CVD4 A0A1L8DEI6 A0A0V0GBF3 D2CG57 A0A182LZQ5 A0A3L8D7U5 A0A194PSH8 U5EWC3 A0A3S2TCP3 A0A2J7RPY4 A0A2W1BMH3 B0XDE9 A0A194PR44 A0A151WZG1 A0A0L7KTQ3 A0A195E542 A0A182SYI8 U5EVF0 A0A182KSD5 A0A067RGL9 A0A026W5E8 A0A023ETG7 A0A212EJ24 A0A3S2L9U7 A0A151WY09 A0A437AVL7 B0XDF0 A0A1Y1KD84 A0A182Y1J3 H9IVN2 E2A7K4 A0A1I8MQZ5 A0A2H1V9D8 A0A084WCB2 Q7Q0C3 A0A212F848 A0A182QUL1 A0A195D353 A0A1B0DMI4 A0A2A4K5Q3 A0A182J645 A0A195ETW6 A0A2A4K6M9 Q2M044 A0A0A1WIH3 W8BAQ3 A0A2W1BRL6
A0A2A4J5Q3 A0A2H1V1L3 A0A2A4J455 A0A194QC06 A0A437B2S9 A0A0N1PI45 A0A3S2L3J0 A0A194QI00 A0A194QC91 A0A2A4JC88 A0A2H1VFS1 A0A194RIS2 A0A3S2NIQ2 A0A3S2M6B8 A0A212F2Q3 A0A212ERF4 A0A212EJ20 A0A1B6D578 A0A2H1V9B3 A0A067RSY2 A0A3S2NA23 A0A2J7RPY7 A0A2J7RPY0 A0A2W1BRJ1 A0A2A4K5R4 A0A2J7RPZ7 A0A2H1VA76 A0A194RF43 A0A2H1W2A4 A0A2H1VDM1 A0A2A4K7E3 A0A2A4JU15 A0A194PT22 A0A182GYP2 A0A0L7RIG1 A0A154PCE9 B0XDF1 Q16WK7 A0A088AAF9 V9ICM7 A0A195DF58 A0A2A3E3H4 A0A067RDE7 A0A2J7RPY5 A0A2A4K5Q2 A0A158NQD1 A0A1L8DEQ3 A0A195CVD4 A0A1L8DEI6 A0A0V0GBF3 D2CG57 A0A182LZQ5 A0A3L8D7U5 A0A194PSH8 U5EWC3 A0A3S2TCP3 A0A2J7RPY4 A0A2W1BMH3 B0XDE9 A0A194PR44 A0A151WZG1 A0A0L7KTQ3 A0A195E542 A0A182SYI8 U5EVF0 A0A182KSD5 A0A067RGL9 A0A026W5E8 A0A023ETG7 A0A212EJ24 A0A3S2L9U7 A0A151WY09 A0A437AVL7 B0XDF0 A0A1Y1KD84 A0A182Y1J3 H9IVN2 E2A7K4 A0A1I8MQZ5 A0A2H1V9D8 A0A084WCB2 Q7Q0C3 A0A212F848 A0A182QUL1 A0A195D353 A0A1B0DMI4 A0A2A4K5Q3 A0A182J645 A0A195ETW6 A0A2A4K6M9 Q2M044 A0A0A1WIH3 W8BAQ3 A0A2W1BRL6
PDB
6N3I
E-value=1.14761e-09,
Score=153
Ontologies
GO
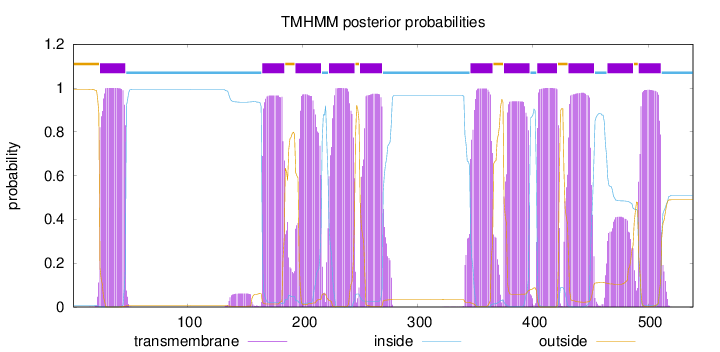
Topology
Length:
539
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
216.86126
Exp number, first 60 AAs:
22.54155
Total prob of N-in:
0.00694
POSSIBLE N-term signal
sequence
outside
1 - 23
TMhelix
24 - 46
inside
47 - 164
TMhelix
165 - 184
outside
185 - 193
TMhelix
194 - 216
inside
217 - 222
TMhelix
223 - 245
outside
246 - 249
TMhelix
250 - 269
inside
270 - 345
TMhelix
346 - 365
outside
366 - 374
TMhelix
375 - 397
inside
398 - 403
TMhelix
404 - 421
outside
422 - 430
TMhelix
431 - 453
inside
454 - 464
TMhelix
465 - 487
outside
488 - 491
TMhelix
492 - 511
inside
512 - 539
Population Genetic Test Statistics
Pi
21.148654
Theta
19.733663
Tajima's D
0.191165
CLR
1.954267
CSRT
0.427378631068447
Interpretation
Uncertain
