Gene
KWMTBOMO12696
Annotation
PREDICTED:_uncharacterized_protein_LOC101746950_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.624
Sequence
CDS
ATGAGAGCCATAATTTTATTATTTTTAATATTCTTTTTTGATTTCCGTGAATGCAGTCACACGTTTGTGGGCACAAGTGTAATGAGACCACTCGTGTACAAGACCAATGCAACCTACAGCGCCCTAACGTTCAGGAAAAGAATAGAATTTATGAACTACACTATTCCAAAAAGCGGAAATTCAGTTATACAGGGCATTCTGGCCTACGATCGGACGCATTCCGGAGCAACCGCCAACATAACTGAAGGAGGAATAGGCTACAACAAAGTCAAGATACGCATGAAGAGCGACAGGGGAAACGAAATCAACTATGAGATTTATATATACGCTTGA
Protein
MRAIILLFLIFFFDFRECSHTFVGTSVMRPLVYKTNATYSALTFRKRIEFMNYTIPKSGNSVIQGILAYDRTHSGATANITEGGIGYNKVKIRMKSDRGNEINYEIYIYA
Summary
Uniprot
O02352
O96053
A0A2A4JML9
A0A3S2P0Z2
A0A0K8SP19
O96054
+ More
A0A1E1WSD0 A0A2H1WFP4 A0A2A4JL79 H9IVV7 H9IVV8 A0A2H1W980 A0A212EUP2 A0A2H1WFP0 S4PFS4 A0A2H1WK39 A0A2A4J1A1 A0A2W1BY77 C1PIJ1 A0A212EJV8 A0A437AZS1 A0A2A4JWT1 H9JA06 A0A1E1WUL5 S4P9G5 A0A437BSZ0 A0A2H1WQ76 A0A212EIW3 A0A194RTL8 A0A194PZC8 A0A0L7KSL9 A0A2W1BYC9 A0A2A4JM91 A0A0L7LMP1 A0A0L7L4F2 A0A2H1VWS1 A0A2H1WDD5 I0B5X1 S4PLY9 S4P6A1 A0A194R3A1 I0B5V1 A0A194PTG4 A0A0N1I7R7 I0B5V2 A0A2W1BSN3 I4DJS8 A0A194PZB4 A0A2H1VX10 A0A194RPM8 H9J189 A0A3S2M8C6 A0A194R4C8 A0A194Q7Z5 A0A1E1WKF6 I4DKH3 I4DJM7 A0A194RCC6 S4PW95 A0A437BQL4 A0A3S2P539 A0A0L7L554 A0A3S2NR50 A0A2W1BMU9 A0A2H1VPR1 A0A2A4IWT7 I0B5V4 A0A194RQ68 A0A194QAE4 A0A0L7KM71 A0A0L7L7T6 A0A0N1IBW3 A0A212EIX0 A0A2A4K508 A0A2W1BUE0 A0A2A4JRI4 A0A2H1VR17 A0A2W1B4F0 A0A212F4T5 H9JW68 A0A182VIK8 A0A182LNW1 A0A182WX71 A0NGU5 A0A182HN34 A0A1E1WVM8 A0A182TP01 A0A182JKE6 A0A182RQP0 A0A2H1WY35 A0A1E1WPV4
A0A1E1WSD0 A0A2H1WFP4 A0A2A4JL79 H9IVV7 H9IVV8 A0A2H1W980 A0A212EUP2 A0A2H1WFP0 S4PFS4 A0A2H1WK39 A0A2A4J1A1 A0A2W1BY77 C1PIJ1 A0A212EJV8 A0A437AZS1 A0A2A4JWT1 H9JA06 A0A1E1WUL5 S4P9G5 A0A437BSZ0 A0A2H1WQ76 A0A212EIW3 A0A194RTL8 A0A194PZC8 A0A0L7KSL9 A0A2W1BYC9 A0A2A4JM91 A0A0L7LMP1 A0A0L7L4F2 A0A2H1VWS1 A0A2H1WDD5 I0B5X1 S4PLY9 S4P6A1 A0A194R3A1 I0B5V1 A0A194PTG4 A0A0N1I7R7 I0B5V2 A0A2W1BSN3 I4DJS8 A0A194PZB4 A0A2H1VX10 A0A194RPM8 H9J189 A0A3S2M8C6 A0A194R4C8 A0A194Q7Z5 A0A1E1WKF6 I4DKH3 I4DJM7 A0A194RCC6 S4PW95 A0A437BQL4 A0A3S2P539 A0A0L7L554 A0A3S2NR50 A0A2W1BMU9 A0A2H1VPR1 A0A2A4IWT7 I0B5V4 A0A194RQ68 A0A194QAE4 A0A0L7KM71 A0A0L7L7T6 A0A0N1IBW3 A0A212EIX0 A0A2A4K508 A0A2W1BUE0 A0A2A4JRI4 A0A2H1VR17 A0A2W1B4F0 A0A212F4T5 H9JW68 A0A182VIK8 A0A182LNW1 A0A182WX71 A0NGU5 A0A182HN34 A0A1E1WVM8 A0A182TP01 A0A182JKE6 A0A182RQP0 A0A2H1WY35 A0A1E1WPV4
Pubmed
EMBL
D70818
BAA20370.1
AB012297
BAA34401.1
NWSH01001065
PCG72834.1
+ More
RSAL01000006 RVE54240.1 GBRD01011274 JAG54550.1 AB012298 BAA34219.1 GDQN01001318 JAT89736.1 ODYU01008370 SOQ51899.1 PCG72835.1 BABH01006586 BABH01006584 BABH01044987 ODYU01006834 SOQ49044.1 AGBW02012353 OWR45174.1 SOQ51898.1 GAIX01002781 JAA89779.1 ODYU01008860 SOQ52824.1 NWSH01004350 PCG65192.1 KZ149899 PZC78605.1 AB499730 BAH57948.1 AGBW02014390 OWR41766.1 RSAL01000239 RVE43687.1 NWSH01000473 PCG76158.1 BABH01009446 BABH01009447 BABH01009448 GDQN01007818 GDQN01000344 JAT83236.1 JAT90710.1 GAIX01009070 JAA83490.1 RSAL01000011 RVE53567.1 ODYU01010243 SOQ55221.1 AGBW02014560 OWR41426.1 KQ459896 KPJ19461.1 KQ459583 KPI98676.1 JTDY01006411 KOB66034.1 PZC78604.1 NWSH01001063 PCG72838.1 JTDY01000603 KOB76496.1 JTDY01003071 KOB70201.1 ODYU01004898 SOQ45248.1 ODYU01007904 SOQ51090.1 JQ660354 AFH57171.1 GAIX01000356 JAA92204.1 GAIX01006931 JAA85629.1 KQ460845 KPJ12004.1 JQ619213 AFH57151.1 KQ459593 KPI96711.1 KQ460940 KPJ10619.1 JQ619214 AFH57152.1 KZ149912 PZC78068.1 AK401546 KQ459592 BAM18168.1 KPI96788.1 KPI98677.1 ODYU01004645 SOQ44744.1 KPJ19462.1 BABH01031461 BABH01031462 RSAL01000010 RVE53781.1 KPJ12085.1 KQ459324 KPJ01662.1 GDQN01003582 JAT87472.1 AK401791 BAM18413.1 AK401495 KQ459603 BAM18117.1 KPI92700.1 KQ460416 KPJ14935.1 GAIX01007178 JAA85382.1 RSAL01000019 RVE52692.1 RSAL01000385 RVE42034.1 JTDY01002960 KOB70409.1 RSAL01000334 RVE42473.1 KZ149959 PZC76392.1 ODYU01003537 SOQ42442.1 NWSH01005557 PCG64129.1 JQ619216 AFH57154.1 KPJ19465.1 KQ459249 KPJ02447.1 JTDY01008778 KOB64403.1 JTDY01002436 KOB71425.1 KQ459992 KPJ18592.1 OWR41429.1 NWSH01000152 PCG78998.1 PZC78609.1 NWSH01000767 PCG74318.1 ODYU01003892 SOQ43216.1 KZ150342 PZC71302.1 AGBW02010320 OWR48732.1 BABH01025361 AAAB01008986 EAU75892.2 APCN01001832 GDQN01009453 GDQN01007785 GDQN01000213 JAT81601.1 JAT83269.1 JAT90841.1 ODYU01011908 SOQ57917.1 GDQN01002024 JAT89030.1
RSAL01000006 RVE54240.1 GBRD01011274 JAG54550.1 AB012298 BAA34219.1 GDQN01001318 JAT89736.1 ODYU01008370 SOQ51899.1 PCG72835.1 BABH01006586 BABH01006584 BABH01044987 ODYU01006834 SOQ49044.1 AGBW02012353 OWR45174.1 SOQ51898.1 GAIX01002781 JAA89779.1 ODYU01008860 SOQ52824.1 NWSH01004350 PCG65192.1 KZ149899 PZC78605.1 AB499730 BAH57948.1 AGBW02014390 OWR41766.1 RSAL01000239 RVE43687.1 NWSH01000473 PCG76158.1 BABH01009446 BABH01009447 BABH01009448 GDQN01007818 GDQN01000344 JAT83236.1 JAT90710.1 GAIX01009070 JAA83490.1 RSAL01000011 RVE53567.1 ODYU01010243 SOQ55221.1 AGBW02014560 OWR41426.1 KQ459896 KPJ19461.1 KQ459583 KPI98676.1 JTDY01006411 KOB66034.1 PZC78604.1 NWSH01001063 PCG72838.1 JTDY01000603 KOB76496.1 JTDY01003071 KOB70201.1 ODYU01004898 SOQ45248.1 ODYU01007904 SOQ51090.1 JQ660354 AFH57171.1 GAIX01000356 JAA92204.1 GAIX01006931 JAA85629.1 KQ460845 KPJ12004.1 JQ619213 AFH57151.1 KQ459593 KPI96711.1 KQ460940 KPJ10619.1 JQ619214 AFH57152.1 KZ149912 PZC78068.1 AK401546 KQ459592 BAM18168.1 KPI96788.1 KPI98677.1 ODYU01004645 SOQ44744.1 KPJ19462.1 BABH01031461 BABH01031462 RSAL01000010 RVE53781.1 KPJ12085.1 KQ459324 KPJ01662.1 GDQN01003582 JAT87472.1 AK401791 BAM18413.1 AK401495 KQ459603 BAM18117.1 KPI92700.1 KQ460416 KPJ14935.1 GAIX01007178 JAA85382.1 RSAL01000019 RVE52692.1 RSAL01000385 RVE42034.1 JTDY01002960 KOB70409.1 RSAL01000334 RVE42473.1 KZ149959 PZC76392.1 ODYU01003537 SOQ42442.1 NWSH01005557 PCG64129.1 JQ619216 AFH57154.1 KPJ19465.1 KQ459249 KPJ02447.1 JTDY01008778 KOB64403.1 JTDY01002436 KOB71425.1 KQ459992 KPJ18592.1 OWR41429.1 NWSH01000152 PCG78998.1 PZC78609.1 NWSH01000767 PCG74318.1 ODYU01003892 SOQ43216.1 KZ150342 PZC71302.1 AGBW02010320 OWR48732.1 BABH01025361 AAAB01008986 EAU75892.2 APCN01001832 GDQN01009453 GDQN01007785 GDQN01000213 JAT81601.1 JAT83269.1 JAT90841.1 ODYU01011908 SOQ57917.1 GDQN01002024 JAT89030.1
Proteomes
Pfam
PF15868 MBF2
Interpro
IPR031734
MBF2
ProteinModelPortal
O02352
O96053
A0A2A4JML9
A0A3S2P0Z2
A0A0K8SP19
O96054
+ More
A0A1E1WSD0 A0A2H1WFP4 A0A2A4JL79 H9IVV7 H9IVV8 A0A2H1W980 A0A212EUP2 A0A2H1WFP0 S4PFS4 A0A2H1WK39 A0A2A4J1A1 A0A2W1BY77 C1PIJ1 A0A212EJV8 A0A437AZS1 A0A2A4JWT1 H9JA06 A0A1E1WUL5 S4P9G5 A0A437BSZ0 A0A2H1WQ76 A0A212EIW3 A0A194RTL8 A0A194PZC8 A0A0L7KSL9 A0A2W1BYC9 A0A2A4JM91 A0A0L7LMP1 A0A0L7L4F2 A0A2H1VWS1 A0A2H1WDD5 I0B5X1 S4PLY9 S4P6A1 A0A194R3A1 I0B5V1 A0A194PTG4 A0A0N1I7R7 I0B5V2 A0A2W1BSN3 I4DJS8 A0A194PZB4 A0A2H1VX10 A0A194RPM8 H9J189 A0A3S2M8C6 A0A194R4C8 A0A194Q7Z5 A0A1E1WKF6 I4DKH3 I4DJM7 A0A194RCC6 S4PW95 A0A437BQL4 A0A3S2P539 A0A0L7L554 A0A3S2NR50 A0A2W1BMU9 A0A2H1VPR1 A0A2A4IWT7 I0B5V4 A0A194RQ68 A0A194QAE4 A0A0L7KM71 A0A0L7L7T6 A0A0N1IBW3 A0A212EIX0 A0A2A4K508 A0A2W1BUE0 A0A2A4JRI4 A0A2H1VR17 A0A2W1B4F0 A0A212F4T5 H9JW68 A0A182VIK8 A0A182LNW1 A0A182WX71 A0NGU5 A0A182HN34 A0A1E1WVM8 A0A182TP01 A0A182JKE6 A0A182RQP0 A0A2H1WY35 A0A1E1WPV4
A0A1E1WSD0 A0A2H1WFP4 A0A2A4JL79 H9IVV7 H9IVV8 A0A2H1W980 A0A212EUP2 A0A2H1WFP0 S4PFS4 A0A2H1WK39 A0A2A4J1A1 A0A2W1BY77 C1PIJ1 A0A212EJV8 A0A437AZS1 A0A2A4JWT1 H9JA06 A0A1E1WUL5 S4P9G5 A0A437BSZ0 A0A2H1WQ76 A0A212EIW3 A0A194RTL8 A0A194PZC8 A0A0L7KSL9 A0A2W1BYC9 A0A2A4JM91 A0A0L7LMP1 A0A0L7L4F2 A0A2H1VWS1 A0A2H1WDD5 I0B5X1 S4PLY9 S4P6A1 A0A194R3A1 I0B5V1 A0A194PTG4 A0A0N1I7R7 I0B5V2 A0A2W1BSN3 I4DJS8 A0A194PZB4 A0A2H1VX10 A0A194RPM8 H9J189 A0A3S2M8C6 A0A194R4C8 A0A194Q7Z5 A0A1E1WKF6 I4DKH3 I4DJM7 A0A194RCC6 S4PW95 A0A437BQL4 A0A3S2P539 A0A0L7L554 A0A3S2NR50 A0A2W1BMU9 A0A2H1VPR1 A0A2A4IWT7 I0B5V4 A0A194RQ68 A0A194QAE4 A0A0L7KM71 A0A0L7L7T6 A0A0N1IBW3 A0A212EIX0 A0A2A4K508 A0A2W1BUE0 A0A2A4JRI4 A0A2H1VR17 A0A2W1B4F0 A0A212F4T5 H9JW68 A0A182VIK8 A0A182LNW1 A0A182WX71 A0NGU5 A0A182HN34 A0A1E1WVM8 A0A182TP01 A0A182JKE6 A0A182RQP0 A0A2H1WY35 A0A1E1WPV4
Ontologies
GO
PANTHER
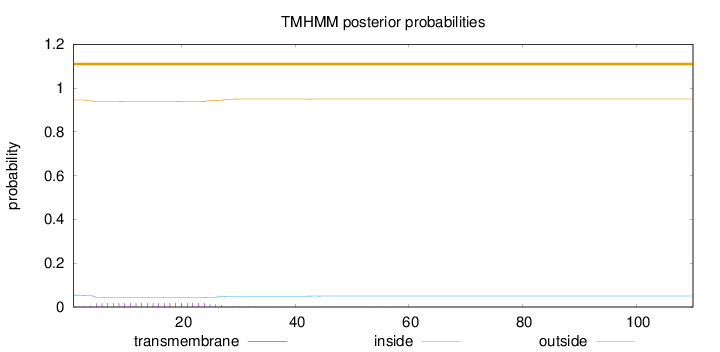
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.971900
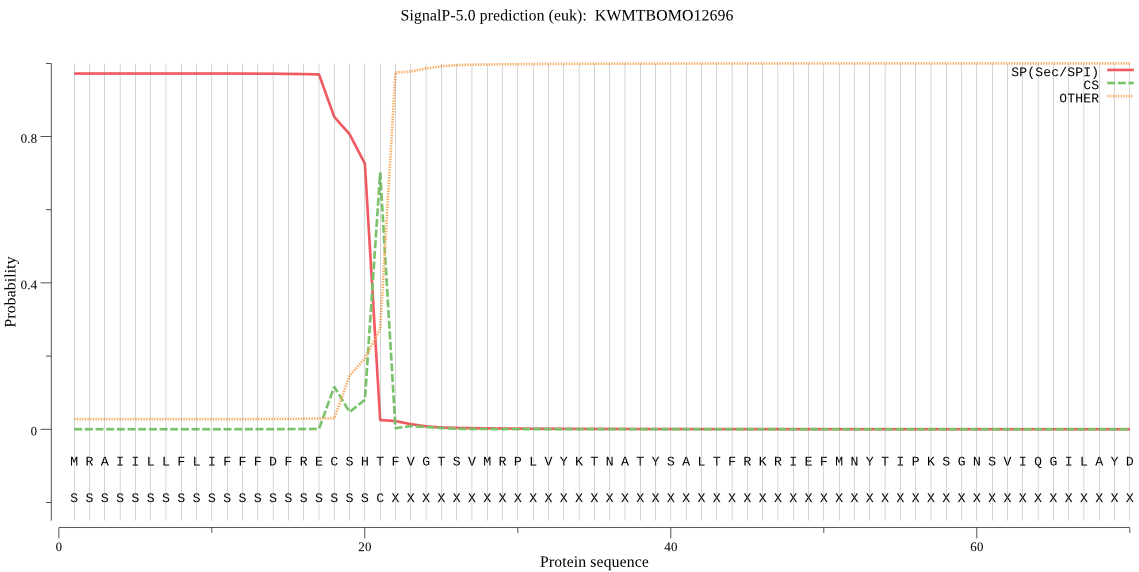
Length:
110
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.41938
Exp number, first 60 AAs:
0.41727
Total prob of N-in:
0.05467
outside
1 - 110
Population Genetic Test Statistics
Pi
19.939756
Theta
19.296442
Tajima's D
0.401972
CLR
0.725245
CSRT
0.486275686215689
Interpretation
Uncertain
