Gene
KWMTBOMO12686 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001595
Annotation
aliphatic_nitrilase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.742
Sequence
CDS
ATGGAGAATGAAACTCACAGCCTTGAATCTATAATAAACAACAACTTGACCGGGCGCGACTTGGAAGAGTTCAACAGAATTCATTTTGGGCGGCGGAATAACCTTGAAATCAAACTGAAAGAATCCTCGATCGCTGCAGCTAAGGAGGCTGACTTCGACGTCGCCGCATACGCTTTCCCGGCCAAGGACGAGCAGACCCGACCCCCGAGAATTGTGAAGGTAGGAATAGTTCAGCATTCCATCGCGGTGCCCACCGATCGTCCAGTCAACGAGCAAAAGAAAGCAATTTTCAATAAAGTCAAGAAAATCATCGATGTTGCCGGCCAAGAGGGTGTTAACATCATCTGTTTCCAAGAGTTGTGGAACATGCCCTTCGCGTTCTGCACAAGGGAGAAGCAGCCGTGGTGCGAGTTTGCCGAATCAGCTGAAGACGGGCCGACCACGACCTTCCTTCGGGAACTCGCCATCAAGTACGCAATGGTGATCGTGTCCTCGATATTAGAAAGGGACGAGAAGCATTCGGACATACTTTGGAACACTGCGGTTGTAATTAGTGACACCGGAAACGTGATCGGGAAACATCGCAAGAACCACATTCCGAGAGTCGGCGATTTTAACGAATCCAACTACTACATGGAAGGTAACACCGGCCATCCTGTATTTGCGACCAGATACGGCAAGATCGCGGTGAACATCTGCTTCGGACGGCACCACGTCTTGAACTGGATGATGTTCGGACAGAACGGAGCGGAGATCGTCTTCAATCCGTCAGCGACGATCGCCGGAGAAGGCGGCAGCGAGTACATGTGGAACGTTGAAGCTAGGAACGCAGCTATCACGAACTGCTACTTCACAGCGGCCATCAACCGAGTCGGCTACGAGGAGTTCCCGAACGAGTTCACTTCGGCTGACGGGAAACCGGCACACAAAGACTTGGGCCTGTTCTACGGATCGAGCTACTTCTGCGGCCCTGACGGCGTTAGGTGCCCCGGTCTGTCCCGCACCAGAGACGGCTTGTTGATCGCGGCCGTAGACTTGAACCTGAACAGACAGATCAAAGACAGACGCTGTTATTACATGACCCAACGCCTGGACATGTACGTGAACAGTCTCAGCAAAGTACTCGAGCTGGATTACAAGCCGCAGGTCGTACACGAAAATGAAAAGCAATTTTAA
Protein
MENETHSLESIINNNLTGRDLEEFNRIHFGRRNNLEIKLKESSIAAAKEADFDVAAYAFPAKDEQTRPPRIVKVGIVQHSIAVPTDRPVNEQKKAIFNKVKKIIDVAGQEGVNIICFQELWNMPFAFCTREKQPWCEFAESAEDGPTTTFLRELAIKYAMVIVSSILERDEKHSDILWNTAVVISDTGNVIGKHRKNHIPRVGDFNESNYYMEGNTGHPVFATRYGKIAVNICFGRHHVLNWMMFGQNGAEIVFNPSATIAGEGGSEYMWNVEARNAAITNCYFTAAINRVGYEEFPNEFTSADGKPAHKDLGLFYGSSYFCGPDGVRCPGLSRTRDGLLIAAVDLNLNRQIKDRRCYYMTQRLDMYVNSLSKVLELDYKPQVVHENEKQF
Summary
Uniprot
H9IWG3
D2Y5R5
A0A2H1VTL9
A0A2A4IWY1
I4DJ34
A0A3G1T199
+ More
A0A194RED5 A0A0L7LGU7 A0A212EYW8 A0A194QCA3 A0A194RE01 A0A0L7KIS7 A0A2H1VLR3 A0A2A4K381 A0A194RPR3 A0A194PYL7 I4DJQ0 S4PAN7 H9IWA5 A0A067R0A3 A0A1J1I3B9 A0A2J7Q1A4 V4ALN2 E9H0A1 A0A0P5BYV4 A0A0N8A500 A0A0P6B4P0 A0A1Q3FMW7 B0W9U3 E0VH43 Q16TC0 A0A336MKH0 A0A0P6J122 A0A336LJV4 A0A3R7PE77 A0A0P5D5E9 A0A1S3IRB8 Q7Q0P4 A0A182I0R9 A0A182JTC6 A0A1L8E2G8 A0A182XM83 A0A182N8R8 A0A1L8E2G6 A0A182P884 A0A1S3J3R0 A0A182Q385 A0A1I8NG00 A0A182VSX3 T1PBW4 A0A182FMW3 B4J8S5 B4MFJ4 A0A1I8PPI7 A0A2L2YEE7 A0A0P4W2K4 U5ETM6 A0A1W4VMA1 A0A0L0BLU0 A0A182YHD8 C3Z990 W8C7U7 B4KRM5 A0A0M4ECJ8 A0A0K8VWP0 A0A0D2VXB6 A0A0A9Z1K6 A0A1J1J3V8 A0A0K8T7L1 B4I4W2 A0A2P8ZES1 A0A182MJX0 B4QZQ2 B3NYQ5 B3LYY1 B4NDC0 A0A1A9Z0N6 A0A0A9Z3V8 Q9VI04 T2M4E1 A0A3S0ZQF3 A0A3Q3A245 A0A1A9YKL3 A0A1B0AM71 B4PRG0 L9JKF3 W5PXC4 Q6NP10 B3RYX4 A0A3Q2FY25 A0A3B3VDT9 B0V1I3 A0A3Q7R8X2 Q7T395 E2QT84 B4IUS1 A0A3P9PRI6 A0A3P9AZ81 A0A2B4S8B2 A0A1Y1HWE1
A0A194RED5 A0A0L7LGU7 A0A212EYW8 A0A194QCA3 A0A194RE01 A0A0L7KIS7 A0A2H1VLR3 A0A2A4K381 A0A194RPR3 A0A194PYL7 I4DJQ0 S4PAN7 H9IWA5 A0A067R0A3 A0A1J1I3B9 A0A2J7Q1A4 V4ALN2 E9H0A1 A0A0P5BYV4 A0A0N8A500 A0A0P6B4P0 A0A1Q3FMW7 B0W9U3 E0VH43 Q16TC0 A0A336MKH0 A0A0P6J122 A0A336LJV4 A0A3R7PE77 A0A0P5D5E9 A0A1S3IRB8 Q7Q0P4 A0A182I0R9 A0A182JTC6 A0A1L8E2G8 A0A182XM83 A0A182N8R8 A0A1L8E2G6 A0A182P884 A0A1S3J3R0 A0A182Q385 A0A1I8NG00 A0A182VSX3 T1PBW4 A0A182FMW3 B4J8S5 B4MFJ4 A0A1I8PPI7 A0A2L2YEE7 A0A0P4W2K4 U5ETM6 A0A1W4VMA1 A0A0L0BLU0 A0A182YHD8 C3Z990 W8C7U7 B4KRM5 A0A0M4ECJ8 A0A0K8VWP0 A0A0D2VXB6 A0A0A9Z1K6 A0A1J1J3V8 A0A0K8T7L1 B4I4W2 A0A2P8ZES1 A0A182MJX0 B4QZQ2 B3NYQ5 B3LYY1 B4NDC0 A0A1A9Z0N6 A0A0A9Z3V8 Q9VI04 T2M4E1 A0A3S0ZQF3 A0A3Q3A245 A0A1A9YKL3 A0A1B0AM71 B4PRG0 L9JKF3 W5PXC4 Q6NP10 B3RYX4 A0A3Q2FY25 A0A3B3VDT9 B0V1I3 A0A3Q7R8X2 Q7T395 E2QT84 B4IUS1 A0A3P9PRI6 A0A3P9AZ81 A0A2B4S8B2 A0A1Y1HWE1
Pubmed
19121390
22651552
26354079
26227816
22118469
23622113
+ More
24845553 23254933 21292972 20566863 17510324 12364791 14747013 17210077 25315136 17994087 26561354 26108605 25244985 18563158 24495485 25401762 26823975 29403074 10731132 11454750 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18336837 26109357 26109356 24065732 17550304 23385571 20809919 18719581 23594743 16341006 25186727 24865297
24845553 23254933 21292972 20566863 17510324 12364791 14747013 17210077 25315136 17994087 26561354 26108605 25244985 18563158 24495485 25401762 26823975 29403074 10731132 11454750 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18336837 26109357 26109356 24065732 17550304 23385571 20809919 18719581 23594743 16341006 25186727 24865297
EMBL
BABH01006545
GU354318
ADB27116.1
ODYU01004363
SOQ44163.1
NWSH01006150
+ More
PCG63633.1 AK401302 KQ459193 BAM17924.1 KPJ03095.1 MG846908 AXY94760.1 KQ460325 KPJ15829.1 JTDY01001176 KOB74647.1 AGBW02011420 OWR46693.1 KPJ03097.1 KPJ15827.1 JTDY01009614 KOB63217.1 ODYU01003259 SOQ41779.1 NWSH01000211 PCG78364.1 KQ459896 KPJ19394.1 KQ459585 KPI98426.1 AK401518 BAM18140.1 GAIX01004921 JAA87639.1 BABH01006272 KK853042 KDR12152.1 CVRI01000037 CRK93358.1 NEVH01019430 PNF22360.1 KB199650 ESP05094.1 GL732580 EFX74852.1 GDIP01181938 JAJ41464.1 GDIP01181937 JAJ41465.1 GDIP01020116 JAM83599.1 GFDL01006163 JAV28882.1 DS231866 EDS40510.1 DS235158 EEB12699.1 CH477653 EAT37751.1 UFQT01001408 SSX30420.1 GDIQ01001384 JAN93353.1 UFQS01000034 UFQT01000034 SSW97906.1 SSX18292.1 QCYY01003537 ROT62833.1 GDIP01162694 JAJ60708.1 AAAB01008980 EAA13946.2 APCN01005251 GFDF01001176 JAV12908.1 GFDF01001177 JAV12907.1 AXCN02001862 KA646257 AFP60886.1 CH916367 EDW02365.1 CH940667 EDW57165.2 IAAA01027348 LAA06508.1 GDRN01077926 GDRN01077925 GDRN01077923 JAI62667.1 GANO01001845 JAB58026.1 JRES01001669 KNC21021.1 GG666599 EEN50819.1 GAMC01003399 JAC03157.1 CH933808 EDW08295.2 CP012524 ALC41465.1 GDHF01009329 JAI42985.1 KE346371 KJE96277.1 GBHO01004492 GDHC01020776 JAG39112.1 JAP97852.1 CVRI01000070 CRL07088.1 GBRD01004313 JAG61508.1 CH480821 EDW55255.1 PYGN01000078 PSN55002.1 AXCM01000430 CM000364 EDX12010.1 CH954181 EDV48168.1 CH902617 EDV41855.1 CH964239 EDW82829.1 GBHO01004491 GBHO01004490 GDHC01017394 JAG39113.1 JAG39114.1 JAQ01235.1 AE014297 AF333187 AAF54141.2 AAK60520.1 HAAD01000553 CDG66785.1 RQTK01000246 RUS83360.1 JXJN01000364 CM000160 EDW96348.1 KB320991 ELW49652.1 AMGL01040103 AMGL01040104 AMGL01040105 AMGL01040106 AMGL01040107 AMGL01040108 BT011121 AAR82788.1 DS985246 EDV23746.1 AL953914 CR848681 BC053204 BC164705 AAH53204.1 AAI64705.1 AAEX03014828 AAEX03014829 AAEX03014830 CH892044 EDX00135.1 LSMT01000146 PFX25616.1 DF237025 GAQ81301.1
PCG63633.1 AK401302 KQ459193 BAM17924.1 KPJ03095.1 MG846908 AXY94760.1 KQ460325 KPJ15829.1 JTDY01001176 KOB74647.1 AGBW02011420 OWR46693.1 KPJ03097.1 KPJ15827.1 JTDY01009614 KOB63217.1 ODYU01003259 SOQ41779.1 NWSH01000211 PCG78364.1 KQ459896 KPJ19394.1 KQ459585 KPI98426.1 AK401518 BAM18140.1 GAIX01004921 JAA87639.1 BABH01006272 KK853042 KDR12152.1 CVRI01000037 CRK93358.1 NEVH01019430 PNF22360.1 KB199650 ESP05094.1 GL732580 EFX74852.1 GDIP01181938 JAJ41464.1 GDIP01181937 JAJ41465.1 GDIP01020116 JAM83599.1 GFDL01006163 JAV28882.1 DS231866 EDS40510.1 DS235158 EEB12699.1 CH477653 EAT37751.1 UFQT01001408 SSX30420.1 GDIQ01001384 JAN93353.1 UFQS01000034 UFQT01000034 SSW97906.1 SSX18292.1 QCYY01003537 ROT62833.1 GDIP01162694 JAJ60708.1 AAAB01008980 EAA13946.2 APCN01005251 GFDF01001176 JAV12908.1 GFDF01001177 JAV12907.1 AXCN02001862 KA646257 AFP60886.1 CH916367 EDW02365.1 CH940667 EDW57165.2 IAAA01027348 LAA06508.1 GDRN01077926 GDRN01077925 GDRN01077923 JAI62667.1 GANO01001845 JAB58026.1 JRES01001669 KNC21021.1 GG666599 EEN50819.1 GAMC01003399 JAC03157.1 CH933808 EDW08295.2 CP012524 ALC41465.1 GDHF01009329 JAI42985.1 KE346371 KJE96277.1 GBHO01004492 GDHC01020776 JAG39112.1 JAP97852.1 CVRI01000070 CRL07088.1 GBRD01004313 JAG61508.1 CH480821 EDW55255.1 PYGN01000078 PSN55002.1 AXCM01000430 CM000364 EDX12010.1 CH954181 EDV48168.1 CH902617 EDV41855.1 CH964239 EDW82829.1 GBHO01004491 GBHO01004490 GDHC01017394 JAG39113.1 JAG39114.1 JAQ01235.1 AE014297 AF333187 AAF54141.2 AAK60520.1 HAAD01000553 CDG66785.1 RQTK01000246 RUS83360.1 JXJN01000364 CM000160 EDW96348.1 KB320991 ELW49652.1 AMGL01040103 AMGL01040104 AMGL01040105 AMGL01040106 AMGL01040107 AMGL01040108 BT011121 AAR82788.1 DS985246 EDV23746.1 AL953914 CR848681 BC053204 BC164705 AAH53204.1 AAI64705.1 AAEX03014828 AAEX03014829 AAEX03014830 CH892044 EDX00135.1 LSMT01000146 PFX25616.1 DF237025 GAQ81301.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000027135 UP000183832 UP000235965 UP000030746 UP000000305 UP000002320 UP000009046 UP000008820 UP000283509 UP000085678 UP000007062 UP000075840 UP000075881 UP000076407 UP000075884 UP000075885 UP000075886 UP000095301 UP000075920 UP000069272 UP000001070 UP000008792 UP000095300 UP000192221 UP000037069 UP000076408 UP000001554 UP000009192 UP000092553 UP000008743 UP000001292 UP000245037 UP000075883 UP000000304 UP000008711 UP000007801 UP000007798 UP000092445 UP000000803 UP000271974 UP000264800 UP000092443 UP000092460 UP000002282 UP000011518 UP000002356 UP000009022 UP000265020 UP000261500 UP000000437 UP000286640 UP000002254 UP000242638 UP000265160 UP000225706 UP000054558
UP000027135 UP000183832 UP000235965 UP000030746 UP000000305 UP000002320 UP000009046 UP000008820 UP000283509 UP000085678 UP000007062 UP000075840 UP000075881 UP000076407 UP000075884 UP000075885 UP000075886 UP000095301 UP000075920 UP000069272 UP000001070 UP000008792 UP000095300 UP000192221 UP000037069 UP000076408 UP000001554 UP000009192 UP000092553 UP000008743 UP000001292 UP000245037 UP000075883 UP000000304 UP000008711 UP000007801 UP000007798 UP000092445 UP000000803 UP000271974 UP000264800 UP000092443 UP000092460 UP000002282 UP000011518 UP000002356 UP000009022 UP000265020 UP000261500 UP000000437 UP000286640 UP000002254 UP000242638 UP000265160 UP000225706 UP000054558
SUPFAM
SSF56317
SSF56317
Gene 3D
ProteinModelPortal
H9IWG3
D2Y5R5
A0A2H1VTL9
A0A2A4IWY1
I4DJ34
A0A3G1T199
+ More
A0A194RED5 A0A0L7LGU7 A0A212EYW8 A0A194QCA3 A0A194RE01 A0A0L7KIS7 A0A2H1VLR3 A0A2A4K381 A0A194RPR3 A0A194PYL7 I4DJQ0 S4PAN7 H9IWA5 A0A067R0A3 A0A1J1I3B9 A0A2J7Q1A4 V4ALN2 E9H0A1 A0A0P5BYV4 A0A0N8A500 A0A0P6B4P0 A0A1Q3FMW7 B0W9U3 E0VH43 Q16TC0 A0A336MKH0 A0A0P6J122 A0A336LJV4 A0A3R7PE77 A0A0P5D5E9 A0A1S3IRB8 Q7Q0P4 A0A182I0R9 A0A182JTC6 A0A1L8E2G8 A0A182XM83 A0A182N8R8 A0A1L8E2G6 A0A182P884 A0A1S3J3R0 A0A182Q385 A0A1I8NG00 A0A182VSX3 T1PBW4 A0A182FMW3 B4J8S5 B4MFJ4 A0A1I8PPI7 A0A2L2YEE7 A0A0P4W2K4 U5ETM6 A0A1W4VMA1 A0A0L0BLU0 A0A182YHD8 C3Z990 W8C7U7 B4KRM5 A0A0M4ECJ8 A0A0K8VWP0 A0A0D2VXB6 A0A0A9Z1K6 A0A1J1J3V8 A0A0K8T7L1 B4I4W2 A0A2P8ZES1 A0A182MJX0 B4QZQ2 B3NYQ5 B3LYY1 B4NDC0 A0A1A9Z0N6 A0A0A9Z3V8 Q9VI04 T2M4E1 A0A3S0ZQF3 A0A3Q3A245 A0A1A9YKL3 A0A1B0AM71 B4PRG0 L9JKF3 W5PXC4 Q6NP10 B3RYX4 A0A3Q2FY25 A0A3B3VDT9 B0V1I3 A0A3Q7R8X2 Q7T395 E2QT84 B4IUS1 A0A3P9PRI6 A0A3P9AZ81 A0A2B4S8B2 A0A1Y1HWE1
A0A194RED5 A0A0L7LGU7 A0A212EYW8 A0A194QCA3 A0A194RE01 A0A0L7KIS7 A0A2H1VLR3 A0A2A4K381 A0A194RPR3 A0A194PYL7 I4DJQ0 S4PAN7 H9IWA5 A0A067R0A3 A0A1J1I3B9 A0A2J7Q1A4 V4ALN2 E9H0A1 A0A0P5BYV4 A0A0N8A500 A0A0P6B4P0 A0A1Q3FMW7 B0W9U3 E0VH43 Q16TC0 A0A336MKH0 A0A0P6J122 A0A336LJV4 A0A3R7PE77 A0A0P5D5E9 A0A1S3IRB8 Q7Q0P4 A0A182I0R9 A0A182JTC6 A0A1L8E2G8 A0A182XM83 A0A182N8R8 A0A1L8E2G6 A0A182P884 A0A1S3J3R0 A0A182Q385 A0A1I8NG00 A0A182VSX3 T1PBW4 A0A182FMW3 B4J8S5 B4MFJ4 A0A1I8PPI7 A0A2L2YEE7 A0A0P4W2K4 U5ETM6 A0A1W4VMA1 A0A0L0BLU0 A0A182YHD8 C3Z990 W8C7U7 B4KRM5 A0A0M4ECJ8 A0A0K8VWP0 A0A0D2VXB6 A0A0A9Z1K6 A0A1J1J3V8 A0A0K8T7L1 B4I4W2 A0A2P8ZES1 A0A182MJX0 B4QZQ2 B3NYQ5 B3LYY1 B4NDC0 A0A1A9Z0N6 A0A0A9Z3V8 Q9VI04 T2M4E1 A0A3S0ZQF3 A0A3Q3A245 A0A1A9YKL3 A0A1B0AM71 B4PRG0 L9JKF3 W5PXC4 Q6NP10 B3RYX4 A0A3Q2FY25 A0A3B3VDT9 B0V1I3 A0A3Q7R8X2 Q7T395 E2QT84 B4IUS1 A0A3P9PRI6 A0A3P9AZ81 A0A2B4S8B2 A0A1Y1HWE1
PDB
2VHI
E-value=1.43203e-129,
Score=1185
Ontologies
PATHWAY
00240
Pyrimidine metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00770 Pantothenate and CoA biosynthesis - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00770 Pantothenate and CoA biosynthesis - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
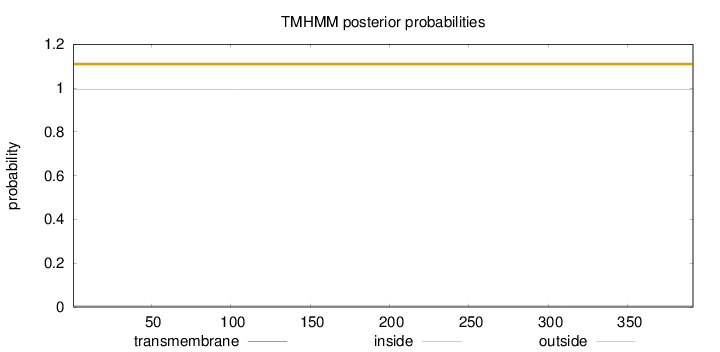
Topology
Length:
391
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00524999999999997
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00644
outside
1 - 391
Population Genetic Test Statistics
Pi
14.715189
Theta
13.096738
Tajima's D
0.286641
CLR
0.44773
CSRT
0.462576871156442
Interpretation
Uncertain
