Gene
KWMTBOMO12678
Pre Gene Modal
BGIBMGA001590
Annotation
PREDICTED:_OTU_domain-containing_protein_7B-like_[Bombyx_mori]
Full name
Proteasome endopeptidase complex
Location in the cell
Nuclear Reliability : 1.794
Sequence
CDS
ATGGTGATGGAAGCAGATCCGATAGCCGAAAAACGAAATGACGAGAGAGTGCACAGTGCGCCTTTGAAGGCAAGGGCGGGCGGTGACGTCATGGGAGAGCGTTCAGTCGATACTTTGGACATGGCCTTCACGGGACCTTCAAATTTAGTGCTCACACCAGCCCCGGAATGTCGGAAACTGTCCCGTGGCATTTCCCGGGCCACAGATAACGAAGGCCTGGTGTGGGCGTTACGTAACAACACAGACCCGGAGCCAAGCACGCTAAGCTCCGATCACATACTGCTCCTACCGGATATAACGGTTTACCCGCCGGATTTCAGAACATTCTTGGAGAAGGACTTACTAGAGACAGCCACTTTGGTCACATTAGAGACGGCAAATATATTGAACTGGTGGTGCCACGGCCGGGTGCCGGGTGCTCCTCGACTGCTGCCACTAGCGACGTCAGGAGACGGGAACTGCTTACCTCATGCGGCGTCATTAGCGGCCTACGGCTTCCACGACAGATTGCTCGCTTTGAGGACCAAAGTACAGGCTCTACTGTCCGGGGAATACGGCGAAACTATAACGAACGCGATCCGGCGTCGCTGGCGCTGGTCTGAGAGCGCGTCGCTGTGCGCGGCGGGGCTGGTCCCGTCGGAGGGCGAGTGGGCGCGCGAGTGGCGCGAGGCGGTGGCGGCGGCGGCCCCCACCCCCCGCGCCCCGCACCCGCAGCACGCGCCGCACTACACCGGGCTGGACGCGCTGCACGTGCTCGCGCTCGCGCACGTCATGCGGCGCCCGGTGCTCGTGTTCGCCGACGTCGCTCTCAGGGATTACCACGGTGACCCGATAGCGCCGATCCCGTTCGGGGGCGTCTATCTGCCGTTGGAACTGGACCCGGCGCAGTGCAGCCGCGCGCCCGTGCTGCTGGCCTACGACGCGGGACACTTCTCCGCGCTGGTCCCCTGCGAGCCCCTGCCCGCCGCCGGGGCCCGAGTGCCGCTCGAAGACAGCGCTGGCAACCCTATGCCGATCAGGTTCAGTGTAGATCCAGGCGAAGACTTCCGCTGGGACGTGGAGCACGATCAAAAAGTTATCAACAACCTCCTCCCGGATGAATACGAGAGATCCGCCATGCTGGCTGCTTATTTAGACCTCGAGAGGGTTGAATGCGTAACGCAGAATCAACCCCCCGAAGAGCTAAGACGATCGCTGGACGCACTGTCGACTAAAAGCTCGAAACAATTAAATTCTGTCGCCAAACAGTTTGGCAGCATCGGCAGGTCTATGAGCAGTAAGATAAAGAAGAATTTCGGGTCCATGGCCAAACTGACTGGCAAGAGCAGCGGCAGTCAAAGTAATCCTGAAGAGAGCTTGGTGCGGAGGCAGTCGACGTGCGAGGTGCTCTGTTGCAGGGTGCTAGCCGCAAGAGCACCTGTGCAAGAAGAAATGGTCAAAAACTATCTCAACGAAGCATGGACGGGGTACACCGCGGAAAAGAACCGCAAGGAAGCGTCGCCCTGCGAGCCCGGGCAGCCGCGGTACGGCACCGGCAAGTCGCAGTTCTACGCGGAGGCGGACCGCGCGGCGCACGAGCTGGCCCGCAGCCACGCCGCGCGCTCGGGCCGCCCCGCCGCCGACCGCACGCTCTACCTGTCCCGGTCAACGTTCTACGTCGACCGCCCGCCCTCCCCGCGCCCCTGCCGCGCGCCCCTCTGCGCCTACTACGGCAGCCCCGCCAACGACGACTACTGCTCGAGGTGTGCGAAACTGCAGTGA
Protein
MVMEADPIAEKRNDERVHSAPLKARAGGDVMGERSVDTLDMAFTGPSNLVLTPAPECRKLSRGISRATDNEGLVWALRNNTDPEPSTLSSDHILLLPDITVYPPDFRTFLEKDLLETATLVTLETANILNWWCHGRVPGAPRLLPLATSGDGNCLPHAASLAAYGFHDRLLALRTKVQALLSGEYGETITNAIRRRWRWSESASLCAAGLVPSEGEWAREWREAVAAAAPTPRAPHPQHAPHYTGLDALHVLALAHVMRRPVLVFADVALRDYHGDPIAPIPFGGVYLPLELDPAQCSRAPVLLAYDAGHFSALVPCEPLPAAGARVPLEDSAGNPMPIRFSVDPGEDFRWDVEHDQKVINNLLPDEYERSAMLAAYLDLERVECVTQNQPPEELRRSLDALSTKSSKQLNSVAKQFGSIGRSMSSKIKKNFGSMAKLTGKSSGSQSNPEESLVRRQSTCEVLCCRVLAARAPVQEEMVKNYLNEAWTGYTAEKNRKEASPCEPGQPRYGTGKSQFYAEADRAAHELARSHAARSGRPAADRTLYLSRSTFYVDRPPSPRPCRAPLCAYYGSPANDDYCSRCAKLQ
Summary
Catalytic Activity
Cleavage of peptide bonds with very broad specificity.
Similarity
Belongs to the peptidase T1A family.
Uniprot
A0A2A4JQ31
A0A212F7A1
A0A2H1VDW9
A0A3S2PKT0
H9IWF8
A0A194QDR2
+ More
A0A194REC3 A0A2H1WSX5 A0A088AV33 A0A2A3ESQ1 A0A0L7R7P8 E2ARU1 F4WE71 A0A154NZA4 A0A0J7P029 A0A151IRT1 A0A151JYJ9 A0A1B6DXN3 A0A1B6ITD5 A0A158NZ45 T1IG62 A0A151I5L0 A0A151XJ64 A0A0M8ZRT6 A0A232ETT1 A0A023F3G1 A0A1B6L9T1 A0A195D1R1 E2BZH6 K7IXF6 A0A026VYT2 A0A3L8D6D3 A0A2R5LNF1 A0A1B6GRG3 A0A1J1HR17 T1IMA8 A0A2J7PM88 A0A2J7PMB6 A0A2P6KZL1 A0A1L8DLE2 A0A2H8TME0 A0A2S2PWK6 J9K7A3 A0A0A9XE76 A0A0K8TA28 A0A146LF34 A0A2M4BD17 U5ESB2 E0VS90 A0A224XD60 K1PEI7 A0A443QVX4 A0A2M4A3G7 A0A2M4A3N8 A0A2M4A3R6 B0XL42 Q177V0 A0A2M4A3H9 A0A1W4WNX4 A0A1W4WYM9 A0A1B6DAP4 A0A182GCR4 A0A0P5C9M8 A0A0P5B4F5 A0A182FBX5 A0A0P6G519 A0A0P5UE29 A0A0P5CUE2 A0A0N8C9Q9 A0A164RH57 A0A0N8CKI4 A0A0P5Z9K0 A0A1D1UIQ2 A0A210QDX1 A0A444SGE9 A0A0A9XWA0 E9HPY1 A0A2C9K1J1 A0A1W4WNX6 A0A433SL36 A0A0P5YNL5 A0A1B6EUU0 V4BIC9 A0A1S3CXX9 A0A3P9KTQ6 A0A3P9ICC1 A0A0B7ACJ6 A0A3Q0SLQ1 C3YQ63 V9KZM8 F6XXD7 A0A443SRW2 U3IG83 A0A3Q3QK01 A0A093BP81
A0A194REC3 A0A2H1WSX5 A0A088AV33 A0A2A3ESQ1 A0A0L7R7P8 E2ARU1 F4WE71 A0A154NZA4 A0A0J7P029 A0A151IRT1 A0A151JYJ9 A0A1B6DXN3 A0A1B6ITD5 A0A158NZ45 T1IG62 A0A151I5L0 A0A151XJ64 A0A0M8ZRT6 A0A232ETT1 A0A023F3G1 A0A1B6L9T1 A0A195D1R1 E2BZH6 K7IXF6 A0A026VYT2 A0A3L8D6D3 A0A2R5LNF1 A0A1B6GRG3 A0A1J1HR17 T1IMA8 A0A2J7PM88 A0A2J7PMB6 A0A2P6KZL1 A0A1L8DLE2 A0A2H8TME0 A0A2S2PWK6 J9K7A3 A0A0A9XE76 A0A0K8TA28 A0A146LF34 A0A2M4BD17 U5ESB2 E0VS90 A0A224XD60 K1PEI7 A0A443QVX4 A0A2M4A3G7 A0A2M4A3N8 A0A2M4A3R6 B0XL42 Q177V0 A0A2M4A3H9 A0A1W4WNX4 A0A1W4WYM9 A0A1B6DAP4 A0A182GCR4 A0A0P5C9M8 A0A0P5B4F5 A0A182FBX5 A0A0P6G519 A0A0P5UE29 A0A0P5CUE2 A0A0N8C9Q9 A0A164RH57 A0A0N8CKI4 A0A0P5Z9K0 A0A1D1UIQ2 A0A210QDX1 A0A444SGE9 A0A0A9XWA0 E9HPY1 A0A2C9K1J1 A0A1W4WNX6 A0A433SL36 A0A0P5YNL5 A0A1B6EUU0 V4BIC9 A0A1S3CXX9 A0A3P9KTQ6 A0A3P9ICC1 A0A0B7ACJ6 A0A3Q0SLQ1 C3YQ63 V9KZM8 F6XXD7 A0A443SRW2 U3IG83 A0A3Q3QK01 A0A093BP81
EC Number
3.4.25.1
Pubmed
EMBL
NWSH01000885
PCG73704.1
AGBW02009897
OWR49610.1
ODYU01002009
SOQ38981.1
+ More
RSAL01000006 RVE54251.1 BABH01006538 BABH01006539 KQ459193 KPJ03105.1 KQ460325 KPJ15819.1 ODYU01010823 SOQ56163.1 KZ288192 PBC34246.1 KQ414638 KOC66859.1 GL442170 EFN63859.1 GL888102 EGI67509.1 KQ434778 KZC04394.1 LBMM01000591 KMQ98005.1 KQ981112 KYN09409.1 KQ981463 KYN41534.1 GEDC01006862 JAS30436.1 GECU01017524 JAS90182.1 ADTU01000044 ACPB03002700 KQ976425 KYM88084.1 KQ982074 KYQ60429.1 KQ435883 KOX69657.1 NNAY01002226 OXU21770.1 GBBI01002849 JAC15863.1 GEBQ01019517 JAT20460.1 KQ976956 KYN06807.1 GL451627 EFN78874.1 AAZX01004609 KK107558 EZA48963.1 QOIP01000012 RLU15992.1 GGLE01006898 MBY11024.1 GECZ01004748 JAS65021.1 CVRI01000009 CRK88657.1 JH431009 NEVH01024418 PNF17450.1 PNF17449.1 MWRG01003277 PRD31763.1 GFDF01006897 JAV07187.1 GFXV01003296 MBW15101.1 GGMS01000673 MBY69876.1 ABLF02036962 GBHO01031950 GBHO01025305 GBHO01020474 GBHO01020473 GBHO01020472 GBHO01020468 JAG11654.1 JAG18299.1 JAG23130.1 JAG23131.1 JAG23132.1 JAG23136.1 GBRD01003427 GBRD01003425 JAG62394.1 GDHC01011861 JAQ06768.1 GGFJ01001577 MBW50718.1 GANO01003328 JAB56543.1 DS235746 EEB16246.1 GFTR01006030 JAW10396.1 JH817065 EKC19988.1 NCKU01003630 RWS07153.1 GGFK01002045 MBW35366.1 GGFK01002044 MBW35365.1 GGFK01002041 MBW35362.1 DS234233 EDS33336.1 CH477371 EAT42418.1 GGFK01002055 MBW35376.1 GEDC01014633 JAS22665.1 JXUM01056332 JXUM01056333 JXUM01056334 KQ561906 KXJ77210.1 GDIP01189820 JAJ33582.1 GDIP01189821 JAJ33581.1 GDIQ01056990 JAN37747.1 GDIP01131529 JAL72185.1 GDIP01166522 JAJ56880.1 GDIQ01100987 JAL50739.1 LRGB01002190 KZS08649.1 GDIP01122775 JAL80939.1 GDIP01046942 JAM56773.1 BDGG01000001 GAU89321.1 NEDP02004063 OWF46954.1 SAUD01037832 RXG52475.1 GBHO01041403 GBHO01025303 GBHO01020476 GBHO01020475 GBHO01020471 GBRD01003428 GBRD01003426 GDHC01021837 JAG02201.1 JAG18301.1 JAG23128.1 JAG23129.1 JAG23133.1 JAG62393.1 JAP96791.1 GL732712 EFX66207.1 RQTK01001555 RUS69853.1 GDIP01055291 JAM48424.1 GECZ01028043 JAS41726.1 KB202719 ESO88369.1 HACG01030820 CEK77685.1 GG666540 EEN57567.1 JW871824 AFP04342.1 AAMC01074463 AAMC01074464 AAMC01074465 AAMC01074466 AAMC01074467 AAMC01074468 AAMC01074469 AAMC01074470 AAMC01074471 NCKV01000562 RWS30286.1 ADON01126987 KN126402 KFU89352.1
RSAL01000006 RVE54251.1 BABH01006538 BABH01006539 KQ459193 KPJ03105.1 KQ460325 KPJ15819.1 ODYU01010823 SOQ56163.1 KZ288192 PBC34246.1 KQ414638 KOC66859.1 GL442170 EFN63859.1 GL888102 EGI67509.1 KQ434778 KZC04394.1 LBMM01000591 KMQ98005.1 KQ981112 KYN09409.1 KQ981463 KYN41534.1 GEDC01006862 JAS30436.1 GECU01017524 JAS90182.1 ADTU01000044 ACPB03002700 KQ976425 KYM88084.1 KQ982074 KYQ60429.1 KQ435883 KOX69657.1 NNAY01002226 OXU21770.1 GBBI01002849 JAC15863.1 GEBQ01019517 JAT20460.1 KQ976956 KYN06807.1 GL451627 EFN78874.1 AAZX01004609 KK107558 EZA48963.1 QOIP01000012 RLU15992.1 GGLE01006898 MBY11024.1 GECZ01004748 JAS65021.1 CVRI01000009 CRK88657.1 JH431009 NEVH01024418 PNF17450.1 PNF17449.1 MWRG01003277 PRD31763.1 GFDF01006897 JAV07187.1 GFXV01003296 MBW15101.1 GGMS01000673 MBY69876.1 ABLF02036962 GBHO01031950 GBHO01025305 GBHO01020474 GBHO01020473 GBHO01020472 GBHO01020468 JAG11654.1 JAG18299.1 JAG23130.1 JAG23131.1 JAG23132.1 JAG23136.1 GBRD01003427 GBRD01003425 JAG62394.1 GDHC01011861 JAQ06768.1 GGFJ01001577 MBW50718.1 GANO01003328 JAB56543.1 DS235746 EEB16246.1 GFTR01006030 JAW10396.1 JH817065 EKC19988.1 NCKU01003630 RWS07153.1 GGFK01002045 MBW35366.1 GGFK01002044 MBW35365.1 GGFK01002041 MBW35362.1 DS234233 EDS33336.1 CH477371 EAT42418.1 GGFK01002055 MBW35376.1 GEDC01014633 JAS22665.1 JXUM01056332 JXUM01056333 JXUM01056334 KQ561906 KXJ77210.1 GDIP01189820 JAJ33582.1 GDIP01189821 JAJ33581.1 GDIQ01056990 JAN37747.1 GDIP01131529 JAL72185.1 GDIP01166522 JAJ56880.1 GDIQ01100987 JAL50739.1 LRGB01002190 KZS08649.1 GDIP01122775 JAL80939.1 GDIP01046942 JAM56773.1 BDGG01000001 GAU89321.1 NEDP02004063 OWF46954.1 SAUD01037832 RXG52475.1 GBHO01041403 GBHO01025303 GBHO01020476 GBHO01020475 GBHO01020471 GBRD01003428 GBRD01003426 GDHC01021837 JAG02201.1 JAG18301.1 JAG23128.1 JAG23129.1 JAG23133.1 JAG62393.1 JAP96791.1 GL732712 EFX66207.1 RQTK01001555 RUS69853.1 GDIP01055291 JAM48424.1 GECZ01028043 JAS41726.1 KB202719 ESO88369.1 HACG01030820 CEK77685.1 GG666540 EEN57567.1 JW871824 AFP04342.1 AAMC01074463 AAMC01074464 AAMC01074465 AAMC01074466 AAMC01074467 AAMC01074468 AAMC01074469 AAMC01074470 AAMC01074471 NCKV01000562 RWS30286.1 ADON01126987 KN126402 KFU89352.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000005204
UP000053268
UP000053240
+ More
UP000005203 UP000242457 UP000053825 UP000000311 UP000007755 UP000076502 UP000036403 UP000078492 UP000078541 UP000005205 UP000015103 UP000078540 UP000075809 UP000053105 UP000215335 UP000078542 UP000008237 UP000002358 UP000053097 UP000279307 UP000183832 UP000235965 UP000007819 UP000009046 UP000005408 UP000285301 UP000002320 UP000008820 UP000192223 UP000069940 UP000249989 UP000069272 UP000076858 UP000186922 UP000242188 UP000288706 UP000000305 UP000076420 UP000271974 UP000030746 UP000079169 UP000265180 UP000265200 UP000261340 UP000001554 UP000008143 UP000288716 UP000016666 UP000261600
UP000005203 UP000242457 UP000053825 UP000000311 UP000007755 UP000076502 UP000036403 UP000078492 UP000078541 UP000005205 UP000015103 UP000078540 UP000075809 UP000053105 UP000215335 UP000078542 UP000008237 UP000002358 UP000053097 UP000279307 UP000183832 UP000235965 UP000007819 UP000009046 UP000005408 UP000285301 UP000002320 UP000008820 UP000192223 UP000069940 UP000249989 UP000069272 UP000076858 UP000186922 UP000242188 UP000288706 UP000000305 UP000076420 UP000271974 UP000030746 UP000079169 UP000265180 UP000265200 UP000261340 UP000001554 UP000008143 UP000288716 UP000016666 UP000261600
PRIDE
Pfam
Interpro
IPR003323
OTU_dom
+ More
IPR002653 Znf_A20
IPR013087 Znf_C2H2_type
IPR011333 SKP1/BTB/POZ_sf
IPR036236 Znf_C2H2_sf
IPR023332 Proteasome_alpha-type
IPR000210 BTB/POZ_dom
IPR001353 Proteasome_sua/b
IPR000426 Proteasome_asu_N
IPR016050 Proteasome_bsu_CS
IPR029055 Ntn_hydrolases_N
IPR009060 UBA-like_sf
IPR038765 Papain-like_cys_pep_sf
IPR033477 Cezanne-2
IPR006671 Cyclin_N
IPR031658 Cyclin_C_2
IPR013763 Cyclin-like
IPR036915 Cyclin-like_sf
IPR002653 Znf_A20
IPR013087 Znf_C2H2_type
IPR011333 SKP1/BTB/POZ_sf
IPR036236 Znf_C2H2_sf
IPR023332 Proteasome_alpha-type
IPR000210 BTB/POZ_dom
IPR001353 Proteasome_sua/b
IPR000426 Proteasome_asu_N
IPR016050 Proteasome_bsu_CS
IPR029055 Ntn_hydrolases_N
IPR009060 UBA-like_sf
IPR038765 Papain-like_cys_pep_sf
IPR033477 Cezanne-2
IPR006671 Cyclin_N
IPR031658 Cyclin_C_2
IPR013763 Cyclin-like
IPR036915 Cyclin-like_sf
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A2A4JQ31
A0A212F7A1
A0A2H1VDW9
A0A3S2PKT0
H9IWF8
A0A194QDR2
+ More
A0A194REC3 A0A2H1WSX5 A0A088AV33 A0A2A3ESQ1 A0A0L7R7P8 E2ARU1 F4WE71 A0A154NZA4 A0A0J7P029 A0A151IRT1 A0A151JYJ9 A0A1B6DXN3 A0A1B6ITD5 A0A158NZ45 T1IG62 A0A151I5L0 A0A151XJ64 A0A0M8ZRT6 A0A232ETT1 A0A023F3G1 A0A1B6L9T1 A0A195D1R1 E2BZH6 K7IXF6 A0A026VYT2 A0A3L8D6D3 A0A2R5LNF1 A0A1B6GRG3 A0A1J1HR17 T1IMA8 A0A2J7PM88 A0A2J7PMB6 A0A2P6KZL1 A0A1L8DLE2 A0A2H8TME0 A0A2S2PWK6 J9K7A3 A0A0A9XE76 A0A0K8TA28 A0A146LF34 A0A2M4BD17 U5ESB2 E0VS90 A0A224XD60 K1PEI7 A0A443QVX4 A0A2M4A3G7 A0A2M4A3N8 A0A2M4A3R6 B0XL42 Q177V0 A0A2M4A3H9 A0A1W4WNX4 A0A1W4WYM9 A0A1B6DAP4 A0A182GCR4 A0A0P5C9M8 A0A0P5B4F5 A0A182FBX5 A0A0P6G519 A0A0P5UE29 A0A0P5CUE2 A0A0N8C9Q9 A0A164RH57 A0A0N8CKI4 A0A0P5Z9K0 A0A1D1UIQ2 A0A210QDX1 A0A444SGE9 A0A0A9XWA0 E9HPY1 A0A2C9K1J1 A0A1W4WNX6 A0A433SL36 A0A0P5YNL5 A0A1B6EUU0 V4BIC9 A0A1S3CXX9 A0A3P9KTQ6 A0A3P9ICC1 A0A0B7ACJ6 A0A3Q0SLQ1 C3YQ63 V9KZM8 F6XXD7 A0A443SRW2 U3IG83 A0A3Q3QK01 A0A093BP81
A0A194REC3 A0A2H1WSX5 A0A088AV33 A0A2A3ESQ1 A0A0L7R7P8 E2ARU1 F4WE71 A0A154NZA4 A0A0J7P029 A0A151IRT1 A0A151JYJ9 A0A1B6DXN3 A0A1B6ITD5 A0A158NZ45 T1IG62 A0A151I5L0 A0A151XJ64 A0A0M8ZRT6 A0A232ETT1 A0A023F3G1 A0A1B6L9T1 A0A195D1R1 E2BZH6 K7IXF6 A0A026VYT2 A0A3L8D6D3 A0A2R5LNF1 A0A1B6GRG3 A0A1J1HR17 T1IMA8 A0A2J7PM88 A0A2J7PMB6 A0A2P6KZL1 A0A1L8DLE2 A0A2H8TME0 A0A2S2PWK6 J9K7A3 A0A0A9XE76 A0A0K8TA28 A0A146LF34 A0A2M4BD17 U5ESB2 E0VS90 A0A224XD60 K1PEI7 A0A443QVX4 A0A2M4A3G7 A0A2M4A3N8 A0A2M4A3R6 B0XL42 Q177V0 A0A2M4A3H9 A0A1W4WNX4 A0A1W4WYM9 A0A1B6DAP4 A0A182GCR4 A0A0P5C9M8 A0A0P5B4F5 A0A182FBX5 A0A0P6G519 A0A0P5UE29 A0A0P5CUE2 A0A0N8C9Q9 A0A164RH57 A0A0N8CKI4 A0A0P5Z9K0 A0A1D1UIQ2 A0A210QDX1 A0A444SGE9 A0A0A9XWA0 E9HPY1 A0A2C9K1J1 A0A1W4WNX6 A0A433SL36 A0A0P5YNL5 A0A1B6EUU0 V4BIC9 A0A1S3CXX9 A0A3P9KTQ6 A0A3P9ICC1 A0A0B7ACJ6 A0A3Q0SLQ1 C3YQ63 V9KZM8 F6XXD7 A0A443SRW2 U3IG83 A0A3Q3QK01 A0A093BP81
PDB
5LRW
E-value=3.36978e-47,
Score=477
Ontologies
GO
PANTHER
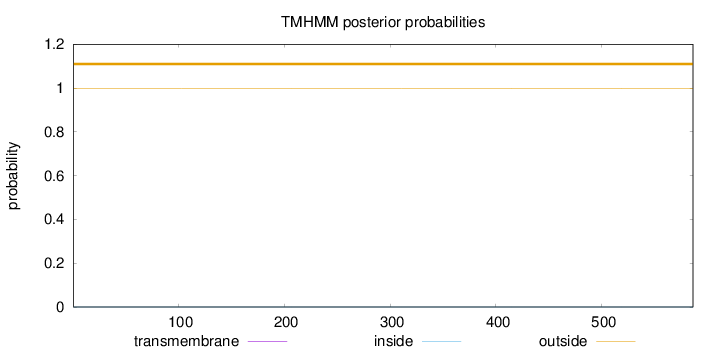
Topology
Subcellular location
Nucleus
Length:
586
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000560000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00050
outside
1 - 586
Population Genetic Test Statistics
Pi
24.951754
Theta
25.13815
Tajima's D
-0.606794
CLR
0.874747
CSRT
0.219489025548723
Interpretation
Uncertain
