Pre Gene Modal
BGIBMGA001581
Annotation
mortality_factor_4-like_[Bombyx_mori]
Full name
Mortality factor 4-like protein 1
Alternative Name
MORF-related gene 15 protein
Transcription factor-like protein MRG15
Transcription factor-like protein MRG15
Location in the cell
Nuclear Reliability : 2.803
Sequence
CDS
ATGGCACCAAAGTTAAAATTTGCTGAAGGTGAAAAAGTGCTATGTTTCCACGGACCGCTAATTTATGAAGCAAAATGTTTAAAAAGCTCAGTTACAAAGGACAAACACGTGAGGTATCTTATTCATTACGCGGGATGGAACAAAAATTGGGATGAATGGGTGCCTGAAAGCCGTGTTCTGAAGTACAATGAAGCAAATGTACAAAGGCAGAAAGAAGTCCAAAGAGCTCATTCCGCACAACCAACTAAAACTAAGAAAACTCCCGCAAAGGGTAGGAAATCTGATGCAGCAGTTGCTGCAACTACTCCAGCTAGAGAAGAATCAAGAGCATCTACCCCTGCGAGTAGTACTCAAGTAAAAGAGTCCGATAGTACACCAGCTCCCGCAAAAACAACTAAAACTCAGAGTAAAGATACACCTGCTGATTCTGGATCTGATCAACCTAAAAAAAAACGAGGACGTCTAGACTTGTCAATAGAATCCGAAGAGCAATACCTAGCAAAAGTAGAAGTTAAGATCAAAATACCCGAAGAACTGAAGGTTTGGCTCGTTGACGACTGGGACGTGATCACAAGGCAACAGAAGTTAGCTATTTTACCAGCAAAATTAACCGTTAGTCAAATAGTGGATAATTATTTGGCTTTCAAGAAATCTAGCAAGTCTCACAACCAGGCTAAGGAGTCTGTGCTGGTCGACATCACGGAGGGGATCAAGGAGTACTTCAACGCGACCCTGGGCTCTCAGCTGCTGTACAAGTTCGAGAGGCCGCAGTACAGCGAGATACTGCAGGAGTATCCCGACACCCCGATGTCTCAGGTGTACGGCGCCGTTCACTTGCTGCGGTTGTTCGCGAAAATGGGTCCGATGCTGGCGTACACGGCGCTCGACGAGAAGTCGCTCCACCACGTGTTGTCGCACATCCAGGACTTCTTGAAGTACATGGTGACGAACAGATCGACGCTTTTCAATTTACAAGATTACGGCAACGCCACGCCTGAATATCATAGGAAAATACAGTAA
Protein
MAPKLKFAEGEKVLCFHGPLIYEAKCLKSSVTKDKHVRYLIHYAGWNKNWDEWVPESRVLKYNEANVQRQKEVQRAHSAQPTKTKKTPAKGRKSDAAVAATTPAREESRASTPASSTQVKESDSTPAPAKTTKTQSKDTPADSGSDQPKKKRGRLDLSIESEEQYLAKVEVKIKIPEELKVWLVDDWDVITRQQKLAILPAKLTVSQIVDNYLAFKKSSKSHNQAKESVLVDITEGIKEYFNATLGSQLLYKFERPQYSEILQEYPDTPMSQVYGAVHLLRLFAKMGPMLAYTALDEKSLHHVLSHIQDFLKYMVTNRSTLFNLQDYGNATPEYHRKIQ
Summary
Description
Component of the NuA4 histone acetyltransferase complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. The NuA4 complex ATPase and helicase activities seem to be, at least in part, contributed by the association of RUVBL1 and RUVBL2 with EP400. NuA4 may also play a direct role in DNA repair when directly recruited to sites of DNA damage. Also component of the mSin3A complex which acts to repress transcription by deacetylation of nucleosomal histones. Required for homologous recombination repair (HRR) and resistance to mitomycin C (MMC). Involved in the localization of PALB2, BRCA2 and RAD51, but not BRCA1, to DNA-damage foci (By similarity).
Subunit
Component of the NuA4 histone acetyltransferase complex which contains the catalytic subunit KAT5/TIP60 and the subunits EP400, TRRAP/PAF400, BRD8/SMAP, EPC1, DMAP1/DNMAP1, RUVBL1/TIP49, RUVBL2, ING3, actin, ACTL6A/BAF53A, MORF4L1/MRG15, MORF4L2/MRGX, MRGBP, YEATS4/GAS41, VPS72/YL1 and MEAF6. The NuA4 complex interacts with MYC. MORF4L1 may also participate in the formation of NuA4 related complexes which lack the KAT5/TIP60 catalytic subunit, but which include the SWI/SNF related protein SRCAP. Component of the mSin3A histone deacetylase complex, which includes SIN3A, HDAC2, ARID4B, MORF4L1, RBBP4/RbAp48, and RBBP7/RbAp46. Interacts with RB1 and MYST1. May also interact with PHF12 and one or more as yet undefined members of the TLE (transducin-like enhancer of split) family of transcriptional repressors. Interacts with the N-terminus of MRFAP1. Found in a complex composed of MORF4L1, MRFAP1 and RB1 Interacts with the entire BRCA complex, which contains BRCA1, PALB2, BRCA2 and RAD51. Interacts with PALB2 (By similarity). Forms a complex with MSL1 and NUPR1 (By similarity).
Keywords
Acetylation
Chromatin regulator
Complete proteome
DNA damage
DNA recombination
DNA repair
Growth regulation
Nucleus
Reference proteome
Transcription
Transcription regulation
Feature
chain Mortality factor 4-like protein 1
Uniprot
C0RWX3
D2CFU1
A0A2H1V4I3
A0A3S2NR87
A0A212F7N0
A0A2A4JLU1
+ More
H9IWF0 A0A1E1W7X3 A0A194RDY0 D6X4Z1 A0A1B0CA64 A0A1Y1K0I5 A0A1L8DX22 A0A1W4WW15 A0A1B0D930 A0A0T6AU28 N6U421 A0A067QMG4 U4UJT0 A0A2J7PYK0 K7INA4 A0A1S4F309 Q17H69 T1JFS7 A0A0K8TKA6 A0A182G1X2 A0A336JXP7 D3TMU9 A0A1B0A4M3 A0A2R5LJP8 A0A1S3JZQ1 A0A023FYY3 L7M4F0 A0A131YRJ9 E2AEL6 A0A3B3CLW0 A0A0N1ITD9 A0A1E1XGG8 Q4SLW2 A0A087ZV33 L7M497 A0A131XC26 E0VRN1 A0A3Q1JH87 A0A3B5BB72 A0A1A8KFU2 A0A1A8BF94 A0A1A8FIU4 A0A3Q3FKW2 A0A1A7XIZ7 A0A1U8DGY5 U3ES96 A0A2D4GVU2 A0A1E1XBF6 A0A131YMM3 A0A3B1JAG6 I3KEC6 A0A218UD67 M3ZN35 A0A147B2Z9 A0A401SXI8 A0A1A8AT52 A0A3B4YIF7 A0A3B4TXQ0 A0A151I2S5 A0A158NB17 H2UNL2 F4WCY1 F1P0X1 H2L9L3 A0A3Q1AIK8 A0A3B3XHV4 A0A226PDY9 A0A3B4UZM9 A0A1A8S1X2 A0A1A8N4U5 V9KRP2 A0A3B4GD74 A0A3Q2X9F4 I3KEC5 A0A087XUK5 A0A3Q3LGK4 Q6AYU1 A0A1V4JDW1 A0A3P8TY52 A0A3B5AXA2 A0A3B3BPL3 A0A3Q2TT47 Q3UBK1 A0A2K6EWU1 A0A2I4BN03 A0A3P9AQW6 A0A3P8PXN9 A0A0F7ZCZ0 A0A0K8RZB4 A0A3P9ILX2 A0A452DY88 A0A2Y9JZY4 K7GQW8
H9IWF0 A0A1E1W7X3 A0A194RDY0 D6X4Z1 A0A1B0CA64 A0A1Y1K0I5 A0A1L8DX22 A0A1W4WW15 A0A1B0D930 A0A0T6AU28 N6U421 A0A067QMG4 U4UJT0 A0A2J7PYK0 K7INA4 A0A1S4F309 Q17H69 T1JFS7 A0A0K8TKA6 A0A182G1X2 A0A336JXP7 D3TMU9 A0A1B0A4M3 A0A2R5LJP8 A0A1S3JZQ1 A0A023FYY3 L7M4F0 A0A131YRJ9 E2AEL6 A0A3B3CLW0 A0A0N1ITD9 A0A1E1XGG8 Q4SLW2 A0A087ZV33 L7M497 A0A131XC26 E0VRN1 A0A3Q1JH87 A0A3B5BB72 A0A1A8KFU2 A0A1A8BF94 A0A1A8FIU4 A0A3Q3FKW2 A0A1A7XIZ7 A0A1U8DGY5 U3ES96 A0A2D4GVU2 A0A1E1XBF6 A0A131YMM3 A0A3B1JAG6 I3KEC6 A0A218UD67 M3ZN35 A0A147B2Z9 A0A401SXI8 A0A1A8AT52 A0A3B4YIF7 A0A3B4TXQ0 A0A151I2S5 A0A158NB17 H2UNL2 F4WCY1 F1P0X1 H2L9L3 A0A3Q1AIK8 A0A3B3XHV4 A0A226PDY9 A0A3B4UZM9 A0A1A8S1X2 A0A1A8N4U5 V9KRP2 A0A3B4GD74 A0A3Q2X9F4 I3KEC5 A0A087XUK5 A0A3Q3LGK4 Q6AYU1 A0A1V4JDW1 A0A3P8TY52 A0A3B5AXA2 A0A3B3BPL3 A0A3Q2TT47 Q3UBK1 A0A2K6EWU1 A0A2I4BN03 A0A3P9AQW6 A0A3P8PXN9 A0A0F7ZCZ0 A0A0K8RZB4 A0A3P9ILX2 A0A452DY88 A0A2Y9JZY4 K7GQW8
Pubmed
22118469
19121390
26354079
18362917
19820115
28004739
+ More
23537049 24845553 20075255 17510324 26369729 26483478 20353571 25576852 26830274 20798317 29451363 28503490 15496914 28049606 20566863 23915248 25329095 25186727 23542700 30297745 21347285 21551351 21719571 15592404 17554307 24621616 24402279 15489334 10349636 11042159 11076861 11217851 12466851 16141073 25727380 30723633
23537049 24845553 20075255 17510324 26369729 26483478 20353571 25576852 26830274 20798317 29451363 28503490 15496914 28049606 20566863 23915248 25329095 25186727 23542700 30297745 21347285 21551351 21719571 15592404 17554307 24621616 24402279 15489334 10349636 11042159 11076861 11217851 12466851 16141073 25727380 30723633
EMBL
DQ888173
ABJ99463.1
DQ888172
ABJ99462.1
ODYU01000608
SOQ35686.1
+ More
RSAL01000006 RVE54257.1 AGBW02009859 OWR49728.1 NWSH01001152 PCG72370.1 BABH01006514 BABH01006515 GDQN01007966 GDQN01001709 JAT83088.1 JAT89345.1 KQ460325 KPJ15807.1 KQ971381 EEZ97599.1 AJWK01003334 GEZM01100705 JAV52816.1 GFDF01003083 JAV11001.1 AJVK01012886 AJVK01012887 LJIG01022808 KRT78610.1 APGK01043667 APGK01043668 KB741015 ENN75371.1 KK853168 KDR10375.1 KB632237 ERL90441.1 NEVH01020346 PNF21421.1 CH477252 EAT46022.1 JH432185 GDAI01002939 JAI14664.1 JXUM01138081 KQ568678 KXJ68922.1 UFQS01000006 UFQT01000006 SSW96981.1 SSX17368.1 CCAG010007751 EZ422751 ADD19027.1 GGLE01005646 MBY09772.1 GBBL01001415 JAC25905.1 GACK01007060 JAA57974.1 GEDV01008036 JAP80521.1 GL438870 EFN68091.1 KQ435830 KOX71724.1 GFAC01000823 JAT98365.1 CAAE01014555 CAF98370.1 GACK01007061 JAA57973.1 GEFH01004669 JAP63912.1 DS235478 EEB16037.1 HAED01015848 HAEE01010484 SBR30534.1 HADZ01001214 HAEA01009000 SBP65155.1 HAEB01012164 HAEC01007701 SBQ58691.1 HADW01016568 HADX01009538 SBP17968.1 GAEP01000943 JAB53878.1 IACJ01150835 LAA63854.1 GFAC01002595 JAT96593.1 GEDV01008034 JAP80523.1 AERX01005350 MUZQ01000419 OWK51616.1 GCES01001169 JAR85154.1 BEZZ01000665 GCC35105.1 HADY01019832 HAEJ01013392 SBP58317.1 KQ976523 KYM82016.1 ADTU01010746 GL888074 EGI67962.1 AADN05000047 AWGT02000098 OXB77607.1 HAEH01020931 HAEI01012749 SBS11553.1 HAEF01013363 HAEG01000857 SBR64110.1 JW868911 AFP01429.1 AYCK01006720 BC078910 LSYS01007908 OPJ70280.1 AK150925 BAE29963.1 GBEX01002041 JAI12519.1 GBKC01002162 GBKD01001302 JAG43908.1 JAG46316.1 LWLT01000019 AEMK02000056 DQIR01208430 DQIR01238113 HDB63907.1
RSAL01000006 RVE54257.1 AGBW02009859 OWR49728.1 NWSH01001152 PCG72370.1 BABH01006514 BABH01006515 GDQN01007966 GDQN01001709 JAT83088.1 JAT89345.1 KQ460325 KPJ15807.1 KQ971381 EEZ97599.1 AJWK01003334 GEZM01100705 JAV52816.1 GFDF01003083 JAV11001.1 AJVK01012886 AJVK01012887 LJIG01022808 KRT78610.1 APGK01043667 APGK01043668 KB741015 ENN75371.1 KK853168 KDR10375.1 KB632237 ERL90441.1 NEVH01020346 PNF21421.1 CH477252 EAT46022.1 JH432185 GDAI01002939 JAI14664.1 JXUM01138081 KQ568678 KXJ68922.1 UFQS01000006 UFQT01000006 SSW96981.1 SSX17368.1 CCAG010007751 EZ422751 ADD19027.1 GGLE01005646 MBY09772.1 GBBL01001415 JAC25905.1 GACK01007060 JAA57974.1 GEDV01008036 JAP80521.1 GL438870 EFN68091.1 KQ435830 KOX71724.1 GFAC01000823 JAT98365.1 CAAE01014555 CAF98370.1 GACK01007061 JAA57973.1 GEFH01004669 JAP63912.1 DS235478 EEB16037.1 HAED01015848 HAEE01010484 SBR30534.1 HADZ01001214 HAEA01009000 SBP65155.1 HAEB01012164 HAEC01007701 SBQ58691.1 HADW01016568 HADX01009538 SBP17968.1 GAEP01000943 JAB53878.1 IACJ01150835 LAA63854.1 GFAC01002595 JAT96593.1 GEDV01008034 JAP80523.1 AERX01005350 MUZQ01000419 OWK51616.1 GCES01001169 JAR85154.1 BEZZ01000665 GCC35105.1 HADY01019832 HAEJ01013392 SBP58317.1 KQ976523 KYM82016.1 ADTU01010746 GL888074 EGI67962.1 AADN05000047 AWGT02000098 OXB77607.1 HAEH01020931 HAEI01012749 SBS11553.1 HAEF01013363 HAEG01000857 SBR64110.1 JW868911 AFP01429.1 AYCK01006720 BC078910 LSYS01007908 OPJ70280.1 AK150925 BAE29963.1 GBEX01002041 JAI12519.1 GBKC01002162 GBKD01001302 JAG43908.1 JAG46316.1 LWLT01000019 AEMK02000056 DQIR01208430 DQIR01238113 HDB63907.1
Proteomes
UP000283053
UP000007151
UP000218220
UP000005204
UP000053240
UP000007266
+ More
UP000092461 UP000192223 UP000092462 UP000019118 UP000027135 UP000030742 UP000235965 UP000002358 UP000008820 UP000069940 UP000249989 UP000092444 UP000092445 UP000085678 UP000000311 UP000261560 UP000053105 UP000007303 UP000005203 UP000009046 UP000265040 UP000261400 UP000261660 UP000189705 UP000018467 UP000005207 UP000197619 UP000002852 UP000265000 UP000287033 UP000261360 UP000261420 UP000078540 UP000005205 UP000005226 UP000007755 UP000000539 UP000001038 UP000257160 UP000261480 UP000198419 UP000261460 UP000264840 UP000028760 UP000261640 UP000002494 UP000190648 UP000265080 UP000233160 UP000192220 UP000265160 UP000265100 UP000265200 UP000291000 UP000248482 UP000008227
UP000092461 UP000192223 UP000092462 UP000019118 UP000027135 UP000030742 UP000235965 UP000002358 UP000008820 UP000069940 UP000249989 UP000092444 UP000092445 UP000085678 UP000000311 UP000261560 UP000053105 UP000007303 UP000005203 UP000009046 UP000265040 UP000261400 UP000261660 UP000189705 UP000018467 UP000005207 UP000197619 UP000002852 UP000265000 UP000287033 UP000261360 UP000261420 UP000078540 UP000005205 UP000005226 UP000007755 UP000000539 UP000001038 UP000257160 UP000261480 UP000198419 UP000261460 UP000264840 UP000028760 UP000261640 UP000002494 UP000190648 UP000265080 UP000233160 UP000192220 UP000265160 UP000265100 UP000265200 UP000291000 UP000248482 UP000008227
Interpro
SUPFAM
SSF54160
SSF54160
Gene 3D
CDD
ProteinModelPortal
C0RWX3
D2CFU1
A0A2H1V4I3
A0A3S2NR87
A0A212F7N0
A0A2A4JLU1
+ More
H9IWF0 A0A1E1W7X3 A0A194RDY0 D6X4Z1 A0A1B0CA64 A0A1Y1K0I5 A0A1L8DX22 A0A1W4WW15 A0A1B0D930 A0A0T6AU28 N6U421 A0A067QMG4 U4UJT0 A0A2J7PYK0 K7INA4 A0A1S4F309 Q17H69 T1JFS7 A0A0K8TKA6 A0A182G1X2 A0A336JXP7 D3TMU9 A0A1B0A4M3 A0A2R5LJP8 A0A1S3JZQ1 A0A023FYY3 L7M4F0 A0A131YRJ9 E2AEL6 A0A3B3CLW0 A0A0N1ITD9 A0A1E1XGG8 Q4SLW2 A0A087ZV33 L7M497 A0A131XC26 E0VRN1 A0A3Q1JH87 A0A3B5BB72 A0A1A8KFU2 A0A1A8BF94 A0A1A8FIU4 A0A3Q3FKW2 A0A1A7XIZ7 A0A1U8DGY5 U3ES96 A0A2D4GVU2 A0A1E1XBF6 A0A131YMM3 A0A3B1JAG6 I3KEC6 A0A218UD67 M3ZN35 A0A147B2Z9 A0A401SXI8 A0A1A8AT52 A0A3B4YIF7 A0A3B4TXQ0 A0A151I2S5 A0A158NB17 H2UNL2 F4WCY1 F1P0X1 H2L9L3 A0A3Q1AIK8 A0A3B3XHV4 A0A226PDY9 A0A3B4UZM9 A0A1A8S1X2 A0A1A8N4U5 V9KRP2 A0A3B4GD74 A0A3Q2X9F4 I3KEC5 A0A087XUK5 A0A3Q3LGK4 Q6AYU1 A0A1V4JDW1 A0A3P8TY52 A0A3B5AXA2 A0A3B3BPL3 A0A3Q2TT47 Q3UBK1 A0A2K6EWU1 A0A2I4BN03 A0A3P9AQW6 A0A3P8PXN9 A0A0F7ZCZ0 A0A0K8RZB4 A0A3P9ILX2 A0A452DY88 A0A2Y9JZY4 K7GQW8
H9IWF0 A0A1E1W7X3 A0A194RDY0 D6X4Z1 A0A1B0CA64 A0A1Y1K0I5 A0A1L8DX22 A0A1W4WW15 A0A1B0D930 A0A0T6AU28 N6U421 A0A067QMG4 U4UJT0 A0A2J7PYK0 K7INA4 A0A1S4F309 Q17H69 T1JFS7 A0A0K8TKA6 A0A182G1X2 A0A336JXP7 D3TMU9 A0A1B0A4M3 A0A2R5LJP8 A0A1S3JZQ1 A0A023FYY3 L7M4F0 A0A131YRJ9 E2AEL6 A0A3B3CLW0 A0A0N1ITD9 A0A1E1XGG8 Q4SLW2 A0A087ZV33 L7M497 A0A131XC26 E0VRN1 A0A3Q1JH87 A0A3B5BB72 A0A1A8KFU2 A0A1A8BF94 A0A1A8FIU4 A0A3Q3FKW2 A0A1A7XIZ7 A0A1U8DGY5 U3ES96 A0A2D4GVU2 A0A1E1XBF6 A0A131YMM3 A0A3B1JAG6 I3KEC6 A0A218UD67 M3ZN35 A0A147B2Z9 A0A401SXI8 A0A1A8AT52 A0A3B4YIF7 A0A3B4TXQ0 A0A151I2S5 A0A158NB17 H2UNL2 F4WCY1 F1P0X1 H2L9L3 A0A3Q1AIK8 A0A3B3XHV4 A0A226PDY9 A0A3B4UZM9 A0A1A8S1X2 A0A1A8N4U5 V9KRP2 A0A3B4GD74 A0A3Q2X9F4 I3KEC5 A0A087XUK5 A0A3Q3LGK4 Q6AYU1 A0A1V4JDW1 A0A3P8TY52 A0A3B5AXA2 A0A3B3BPL3 A0A3Q2TT47 Q3UBK1 A0A2K6EWU1 A0A2I4BN03 A0A3P9AQW6 A0A3P8PXN9 A0A0F7ZCZ0 A0A0K8RZB4 A0A3P9ILX2 A0A452DY88 A0A2Y9JZY4 K7GQW8
PDB
2AQL
E-value=3.7523e-54,
Score=534
Ontologies
GO
PANTHER
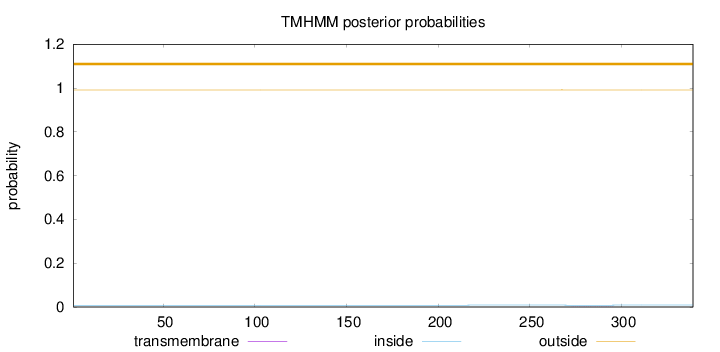
Topology
Subcellular location
Nucleus
Length:
339
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01062
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.00854
outside
1 - 339
Population Genetic Test Statistics
Pi
6.431593
Theta
13.953894
Tajima's D
-1.783294
CLR
1.092956
CSRT
0.0263486825658717
Interpretation
Uncertain
