Gene
KWMTBOMO12670
Pre Gene Modal
BGIBMGA001401
Annotation
PREDICTED:_NAD-dependent_protein_deacetylase_Sirt6_[Plutella_xylostella]
Full name
NAD-dependent protein deacetylase Sirt6
Alternative Name
Regulatory protein SIR2 homolog 6
SIR2-like protein 6
SIR2-like protein 6
Location in the cell
Cytoplasmic Reliability : 2.277 Nuclear Reliability : 1.749
Sequence
CDS
ATGTCTTGCAATTATGCAGAAGGACTATCACCTTATGAAAACAAAGGTATACTAGGAGTCCCAGAGAAATTTGATTCAAATGACAAACTGAACCAGAAATGTGTACTCTTGGCTCAACTTGTGAAAGATAGTAAACATATTGTTGTCCACACTGGAGCTGGCATAAGTACTTCTGCGGGTATTCCAGACTTTAGAGGGCCAAATGGAGTGTGGACTTTAGAGAAGGAAGGTAAAAAGCCAACCATCAATGTATCATTTGCTGATGCACAACCCACAAAAACCCATATGATTTTAAAAAAGTTGGTTGAAATAAAAAAACTTAAATACATTGTGAGTCAAAACATTGACGGACTTCATCTCAAGTCTGGTGTCCCAAGAAAATTCTTAGCTGAACTACATGGAAATATGTTTATTGATGAATGTAACATCTGCAAGAGACAGTTTGTCCGTAGCAGTCCTGTCGAAACTGTAGGCAAGAAATGCAGTGGTGTGCCTTGTGCAGCTCACCATGTGACGGGTAGACCTTGCCGTGGTAGATTATATGACGGAGTGCTTGACTGGGAGCATAGCTTGCCTGAAAATGATTTGCTAATGGCAGAATGGCATTCAAGTATTGCAGACTTGAGCATATGTCTAGGTACAACACTTCAAATAGTACCAAGTGGTAATTTACCACTAGAGACTATTAAATATGGTGGAAAGTTAGTTATTTGCAACCTACAACCAACAAAACATGACAATAAAGCTGATCTCCTAATAAATTATTATGTAGATGATGTACTTGAGAAAGTAATGGACATCTTAGGAATTGAAATCCCCAGTTATAATGAATCTGAGAACCCCATGAAATTTGCAGAAACAGCTATTATTGACTGGTCAATAGACAGAAAAGATGTCTTAGCTTTAGAAAAGACATTCAAATCAAAATGCAAAGGAGTTAAAAAGAAACGAATATTAATAAAAACAAAAAGATTTACCAGTAATGCCAATGATATTGAAAAATCAAAAATGATCAAATTAGAAGTTAAGGAAGAGAATGAAACTAAAGACTGGCCATTGAAACCTTCGGATCAAGAATTTATAATTGATCAAAAGGGTTCTATTGTGTCTTGA
Protein
MSCNYAEGLSPYENKGILGVPEKFDSNDKLNQKCVLLAQLVKDSKHIVVHTGAGISTSAGIPDFRGPNGVWTLEKEGKKPTINVSFADAQPTKTHMILKKLVEIKKLKYIVSQNIDGLHLKSGVPRKFLAELHGNMFIDECNICKRQFVRSSPVETVGKKCSGVPCAAHHVTGRPCRGRLYDGVLDWEHSLPENDLLMAEWHSSIADLSICLGTTLQIVPSGNLPLETIKYGGKLVICNLQPTKHDNKADLLINYYVDDVLEKVMDILGIEIPSYNESENPMKFAETAIIDWSIDRKDVLALEKTFKSKCKGVKKKRILIKTKRFTSNANDIEKSKMIKLEVKEENETKDWPLKPSDQEFIIDQKGSIVS
Summary
Description
NAD-dependent protein deacetylase (By similarity). May be involved in the regulation of life span.
Catalytic Activity
H2O + N(6)-acetyl-L-lysyl-[protein] + NAD(+) = 2''-O-acetyl-ADP-D-ribose + L-lysyl-[protein] + nicotinamide
Cofactor
Zn(2+)
Similarity
Belongs to the sirtuin family. Class IV subfamily.
Belongs to the universal ribosomal protein uL22 family.
Belongs to the universal ribosomal protein uL22 family.
Keywords
Complete proteome
Hydrolase
Metal-binding
NAD
Reference proteome
Zinc
Feature
chain NAD-dependent protein deacetylase Sirt6
Uniprot
H9IVX0
A0A2A4IZK5
A0A2H1VR05
A0A212F7K9
A0A194REA8
A0A194QCC3
+ More
A0A437BUW4 D6W9J7 K7IVH1 E2BAH7 A0A2J7PC12 A0A2J7PC17 A0A154PCL6 A0A0P4VU52 B4JPZ6 A0A0L7R8F1 A0A195EQL6 A0A088AIY8 A0A158NQL6 A0A026W6E6 A0A151XAX5 A0A1I8P496 B4LVC5 F4WKH3 A0A1B6GGY3 A0A1B6L1N7 A0A195BR04 A0A1B0EZ10 A0A347ZJH1 A0A195DI37 A0A1Y9IVF5 B4MUC7 A0A1B6K2Q4 A0A1B6I1F0 E2AN21 A0A1Y1KC95 B4KHR8 A0A1B0CHX5 A0A0R3NRG2 A0A023F1X0 B3LW04 A0A182TFJ6 A0A1C7CZT1 A0A1I8JVJ4 A0A1I8JTF3 Q7PXL0 A0A182LW30 A0A1I8JW60 A0A1Y9HEZ3 A0A2A3E4V8 A0A0L0C8M7 A0A232EZW5 B3P1P7 A0A0A1XA58 Q16PZ4 A0A1B6CB19 A0A1S4FTB4 A0A182S7X0 A0A1Y9H9L0 A0A240PPS8 A0A224XN22 A0A1W4UK10 B4PKN2 A0A1I8NCI0 A0A0K8USI3 A0A069DSU1 A0A1Y9G9J4 A0A034VXN7 A0A240PMX2 Q9VH08 B4HJ75 A0A0M4E8J5 A0A182YGA4 A0A0V0G6Z7 A0A3B0JHK7 A0A1Y9H398 A0A336LIU6 A0A336K0Y7 W5J661 W8AG47 B4QV96 A0A3R7Q068 A0A0K8TLQ7 E0VRL7 A0A1A9VUJ7 A0A1A9YMF0 A0A1A9W8Q5 A0A1A9ZKS0 A0A1B0BDH8 A0A1B0G8R3 A0A2R7VWG6 N6UN31 U4UFK1 A0A3B0JH01 U4TUL2 A0A0A9Z7B5 N6TUQ0 A0A0A9ZCQ7 A0A0A9Z9M5 A0A0P4W8L8
A0A437BUW4 D6W9J7 K7IVH1 E2BAH7 A0A2J7PC12 A0A2J7PC17 A0A154PCL6 A0A0P4VU52 B4JPZ6 A0A0L7R8F1 A0A195EQL6 A0A088AIY8 A0A158NQL6 A0A026W6E6 A0A151XAX5 A0A1I8P496 B4LVC5 F4WKH3 A0A1B6GGY3 A0A1B6L1N7 A0A195BR04 A0A1B0EZ10 A0A347ZJH1 A0A195DI37 A0A1Y9IVF5 B4MUC7 A0A1B6K2Q4 A0A1B6I1F0 E2AN21 A0A1Y1KC95 B4KHR8 A0A1B0CHX5 A0A0R3NRG2 A0A023F1X0 B3LW04 A0A182TFJ6 A0A1C7CZT1 A0A1I8JVJ4 A0A1I8JTF3 Q7PXL0 A0A182LW30 A0A1I8JW60 A0A1Y9HEZ3 A0A2A3E4V8 A0A0L0C8M7 A0A232EZW5 B3P1P7 A0A0A1XA58 Q16PZ4 A0A1B6CB19 A0A1S4FTB4 A0A182S7X0 A0A1Y9H9L0 A0A240PPS8 A0A224XN22 A0A1W4UK10 B4PKN2 A0A1I8NCI0 A0A0K8USI3 A0A069DSU1 A0A1Y9G9J4 A0A034VXN7 A0A240PMX2 Q9VH08 B4HJ75 A0A0M4E8J5 A0A182YGA4 A0A0V0G6Z7 A0A3B0JHK7 A0A1Y9H398 A0A336LIU6 A0A336K0Y7 W5J661 W8AG47 B4QV96 A0A3R7Q068 A0A0K8TLQ7 E0VRL7 A0A1A9VUJ7 A0A1A9YMF0 A0A1A9W8Q5 A0A1A9ZKS0 A0A1B0BDH8 A0A1B0G8R3 A0A2R7VWG6 N6UN31 U4UFK1 A0A3B0JH01 U4TUL2 A0A0A9Z7B5 N6TUQ0 A0A0A9ZCQ7 A0A0A9Z9M5 A0A0P4W8L8
EC Number
3.5.1.-
Pubmed
19121390
22118469
26354079
18362917
19820115
20075255
+ More
20798317 27129103 17994087 21347285 24508170 30249741 21719571 28004739 15632085 25474469 20966253 12364791 14747013 17210077 26108605 28648823 25830018 17510324 17550304 25315136 26334808 25348373 10731132 12537572 17159295 25244985 20920257 23761445 24495485 26369729 20566863 23537049 25401762
20798317 27129103 17994087 21347285 24508170 30249741 21719571 28004739 15632085 25474469 20966253 12364791 14747013 17210077 26108605 28648823 25830018 17510324 17550304 25315136 26334808 25348373 10731132 12537572 17159295 25244985 20920257 23761445 24495485 26369729 20566863 23537049 25401762
EMBL
BABH01006508
BABH01006509
BABH01006510
NWSH01004755
PCG64704.1
ODYU01003897
+ More
SOQ43227.1 AGBW02009859 OWR49724.1 KQ460325 KPJ15804.1 KQ459193 KPJ03117.1 RSAL01000006 RVE54262.1 KQ971312 EEZ98502.1 GL446730 EFN87309.1 NEVH01027073 PNF13869.1 PNF13871.1 KQ434874 KZC09669.1 GDKW01003252 JAI53343.1 CH916372 EDV98976.1 KQ414632 KOC67162.1 KQ982021 KYN30570.1 ADTU01023390 KK107405 QOIP01000014 EZA51196.1 RLU14917.1 KQ982335 KYQ57531.1 CH940649 EDW63304.1 GL888199 EGI65311.1 GECZ01008067 JAS61702.1 GEBQ01022355 JAT17622.1 KQ976419 KYM89286.1 AJVK01007422 FX985739 BBA84477.1 KQ980824 KYN12506.1 CH963857 EDW76053.1 GECU01001954 JAT05753.1 GECU01036617 GECU01026964 GECU01022868 GECU01006688 JAS71089.1 JAS80742.1 JAS84838.1 JAT01019.1 GL441034 EFN65179.1 GEZM01088551 JAV58108.1 CH933807 EDW13355.1 AJWK01012834 AJWK01012835 CH379059 KRT03758.1 GBBI01003175 JAC15537.1 CH902617 EDV41537.1 APCN01000875 AAAB01008987 EAA00841.3 AXCM01005015 KZ288372 PBC26730.1 JRES01000749 KNC28793.1 NNAY01001524 OXU23698.1 CH954181 EDV49646.1 GBXI01006295 JAD07997.1 CH477768 EAT36439.1 GEDC01026664 JAS10634.1 AXCN02000207 GFTR01005218 JAW11208.1 CM000160 EDW96791.1 GDHF01022846 JAI29468.1 GBGD01002098 JAC86791.1 GAKP01012659 JAC46293.1 AE014297 BT126234 CH480815 EDW42747.1 CP012523 ALC39277.1 GECL01002644 JAP03480.1 OUUW01000006 SPP81675.1 UFQS01000011 UFQT01000011 SSW97149.1 SSX17535.1 SSW97148.1 SSX17534.1 ADMH02002026 ETN59902.1 GAMC01019275 JAB87280.1 CM000364 EDX13502.1 MG491309 QCYY01003131 AZI71012.1 ROT65169.1 GDAI01002306 JAI15297.1 DS235478 EEB16023.1 JXJN01012501 CCAG010000303 KK854098 PTY11241.1 APGK01016464 KB739858 ENN82101.1 KB632093 ERL88695.1 SPP81674.1 KB630484 ERL83623.1 GBHO01002507 JAG41097.1 APGK01017821 KB740053 ENN81803.1 GBHO01002506 JAG41098.1 GBHO01002505 JAG41099.1 GDRN01081144 JAI62065.1
SOQ43227.1 AGBW02009859 OWR49724.1 KQ460325 KPJ15804.1 KQ459193 KPJ03117.1 RSAL01000006 RVE54262.1 KQ971312 EEZ98502.1 GL446730 EFN87309.1 NEVH01027073 PNF13869.1 PNF13871.1 KQ434874 KZC09669.1 GDKW01003252 JAI53343.1 CH916372 EDV98976.1 KQ414632 KOC67162.1 KQ982021 KYN30570.1 ADTU01023390 KK107405 QOIP01000014 EZA51196.1 RLU14917.1 KQ982335 KYQ57531.1 CH940649 EDW63304.1 GL888199 EGI65311.1 GECZ01008067 JAS61702.1 GEBQ01022355 JAT17622.1 KQ976419 KYM89286.1 AJVK01007422 FX985739 BBA84477.1 KQ980824 KYN12506.1 CH963857 EDW76053.1 GECU01001954 JAT05753.1 GECU01036617 GECU01026964 GECU01022868 GECU01006688 JAS71089.1 JAS80742.1 JAS84838.1 JAT01019.1 GL441034 EFN65179.1 GEZM01088551 JAV58108.1 CH933807 EDW13355.1 AJWK01012834 AJWK01012835 CH379059 KRT03758.1 GBBI01003175 JAC15537.1 CH902617 EDV41537.1 APCN01000875 AAAB01008987 EAA00841.3 AXCM01005015 KZ288372 PBC26730.1 JRES01000749 KNC28793.1 NNAY01001524 OXU23698.1 CH954181 EDV49646.1 GBXI01006295 JAD07997.1 CH477768 EAT36439.1 GEDC01026664 JAS10634.1 AXCN02000207 GFTR01005218 JAW11208.1 CM000160 EDW96791.1 GDHF01022846 JAI29468.1 GBGD01002098 JAC86791.1 GAKP01012659 JAC46293.1 AE014297 BT126234 CH480815 EDW42747.1 CP012523 ALC39277.1 GECL01002644 JAP03480.1 OUUW01000006 SPP81675.1 UFQS01000011 UFQT01000011 SSW97149.1 SSX17535.1 SSW97148.1 SSX17534.1 ADMH02002026 ETN59902.1 GAMC01019275 JAB87280.1 CM000364 EDX13502.1 MG491309 QCYY01003131 AZI71012.1 ROT65169.1 GDAI01002306 JAI15297.1 DS235478 EEB16023.1 JXJN01012501 CCAG010000303 KK854098 PTY11241.1 APGK01016464 KB739858 ENN82101.1 KB632093 ERL88695.1 SPP81674.1 KB630484 ERL83623.1 GBHO01002507 JAG41097.1 APGK01017821 KB740053 ENN81803.1 GBHO01002506 JAG41098.1 GBHO01002505 JAG41099.1 GDRN01081144 JAI62065.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000007266 UP000002358 UP000008237 UP000235965 UP000076502 UP000001070 UP000053825 UP000078541 UP000005203 UP000005205 UP000053097 UP000279307 UP000075809 UP000095300 UP000008792 UP000007755 UP000078540 UP000092462 UP000078492 UP000075920 UP000007798 UP000000311 UP000009192 UP000092461 UP000001819 UP000007801 UP000075902 UP000075882 UP000075903 UP000075840 UP000007062 UP000075883 UP000076407 UP000075900 UP000242457 UP000037069 UP000215335 UP000008711 UP000008820 UP000075901 UP000075886 UP000075885 UP000192221 UP000002282 UP000095301 UP000069272 UP000075881 UP000000803 UP000001292 UP000092553 UP000076408 UP000268350 UP000075884 UP000000673 UP000000304 UP000283509 UP000009046 UP000078200 UP000092443 UP000091820 UP000092445 UP000092460 UP000092444 UP000019118 UP000030742
UP000007266 UP000002358 UP000008237 UP000235965 UP000076502 UP000001070 UP000053825 UP000078541 UP000005203 UP000005205 UP000053097 UP000279307 UP000075809 UP000095300 UP000008792 UP000007755 UP000078540 UP000092462 UP000078492 UP000075920 UP000007798 UP000000311 UP000009192 UP000092461 UP000001819 UP000007801 UP000075902 UP000075882 UP000075903 UP000075840 UP000007062 UP000075883 UP000076407 UP000075900 UP000242457 UP000037069 UP000215335 UP000008711 UP000008820 UP000075901 UP000075886 UP000075885 UP000192221 UP000002282 UP000095301 UP000069272 UP000075881 UP000000803 UP000001292 UP000092553 UP000076408 UP000268350 UP000075884 UP000000673 UP000000304 UP000283509 UP000009046 UP000078200 UP000092443 UP000091820 UP000092445 UP000092460 UP000092444 UP000019118 UP000030742
Interpro
Gene 3D
ProteinModelPortal
H9IVX0
A0A2A4IZK5
A0A2H1VR05
A0A212F7K9
A0A194REA8
A0A194QCC3
+ More
A0A437BUW4 D6W9J7 K7IVH1 E2BAH7 A0A2J7PC12 A0A2J7PC17 A0A154PCL6 A0A0P4VU52 B4JPZ6 A0A0L7R8F1 A0A195EQL6 A0A088AIY8 A0A158NQL6 A0A026W6E6 A0A151XAX5 A0A1I8P496 B4LVC5 F4WKH3 A0A1B6GGY3 A0A1B6L1N7 A0A195BR04 A0A1B0EZ10 A0A347ZJH1 A0A195DI37 A0A1Y9IVF5 B4MUC7 A0A1B6K2Q4 A0A1B6I1F0 E2AN21 A0A1Y1KC95 B4KHR8 A0A1B0CHX5 A0A0R3NRG2 A0A023F1X0 B3LW04 A0A182TFJ6 A0A1C7CZT1 A0A1I8JVJ4 A0A1I8JTF3 Q7PXL0 A0A182LW30 A0A1I8JW60 A0A1Y9HEZ3 A0A2A3E4V8 A0A0L0C8M7 A0A232EZW5 B3P1P7 A0A0A1XA58 Q16PZ4 A0A1B6CB19 A0A1S4FTB4 A0A182S7X0 A0A1Y9H9L0 A0A240PPS8 A0A224XN22 A0A1W4UK10 B4PKN2 A0A1I8NCI0 A0A0K8USI3 A0A069DSU1 A0A1Y9G9J4 A0A034VXN7 A0A240PMX2 Q9VH08 B4HJ75 A0A0M4E8J5 A0A182YGA4 A0A0V0G6Z7 A0A3B0JHK7 A0A1Y9H398 A0A336LIU6 A0A336K0Y7 W5J661 W8AG47 B4QV96 A0A3R7Q068 A0A0K8TLQ7 E0VRL7 A0A1A9VUJ7 A0A1A9YMF0 A0A1A9W8Q5 A0A1A9ZKS0 A0A1B0BDH8 A0A1B0G8R3 A0A2R7VWG6 N6UN31 U4UFK1 A0A3B0JH01 U4TUL2 A0A0A9Z7B5 N6TUQ0 A0A0A9ZCQ7 A0A0A9Z9M5 A0A0P4W8L8
A0A437BUW4 D6W9J7 K7IVH1 E2BAH7 A0A2J7PC12 A0A2J7PC17 A0A154PCL6 A0A0P4VU52 B4JPZ6 A0A0L7R8F1 A0A195EQL6 A0A088AIY8 A0A158NQL6 A0A026W6E6 A0A151XAX5 A0A1I8P496 B4LVC5 F4WKH3 A0A1B6GGY3 A0A1B6L1N7 A0A195BR04 A0A1B0EZ10 A0A347ZJH1 A0A195DI37 A0A1Y9IVF5 B4MUC7 A0A1B6K2Q4 A0A1B6I1F0 E2AN21 A0A1Y1KC95 B4KHR8 A0A1B0CHX5 A0A0R3NRG2 A0A023F1X0 B3LW04 A0A182TFJ6 A0A1C7CZT1 A0A1I8JVJ4 A0A1I8JTF3 Q7PXL0 A0A182LW30 A0A1I8JW60 A0A1Y9HEZ3 A0A2A3E4V8 A0A0L0C8M7 A0A232EZW5 B3P1P7 A0A0A1XA58 Q16PZ4 A0A1B6CB19 A0A1S4FTB4 A0A182S7X0 A0A1Y9H9L0 A0A240PPS8 A0A224XN22 A0A1W4UK10 B4PKN2 A0A1I8NCI0 A0A0K8USI3 A0A069DSU1 A0A1Y9G9J4 A0A034VXN7 A0A240PMX2 Q9VH08 B4HJ75 A0A0M4E8J5 A0A182YGA4 A0A0V0G6Z7 A0A3B0JHK7 A0A1Y9H398 A0A336LIU6 A0A336K0Y7 W5J661 W8AG47 B4QV96 A0A3R7Q068 A0A0K8TLQ7 E0VRL7 A0A1A9VUJ7 A0A1A9YMF0 A0A1A9W8Q5 A0A1A9ZKS0 A0A1B0BDH8 A0A1B0G8R3 A0A2R7VWG6 N6UN31 U4UFK1 A0A3B0JH01 U4TUL2 A0A0A9Z7B5 N6TUQ0 A0A0A9ZCQ7 A0A0A9Z9M5 A0A0P4W8L8
PDB
3ZG6
E-value=9.61199e-86,
Score=807
Ontologies
GO
Topology
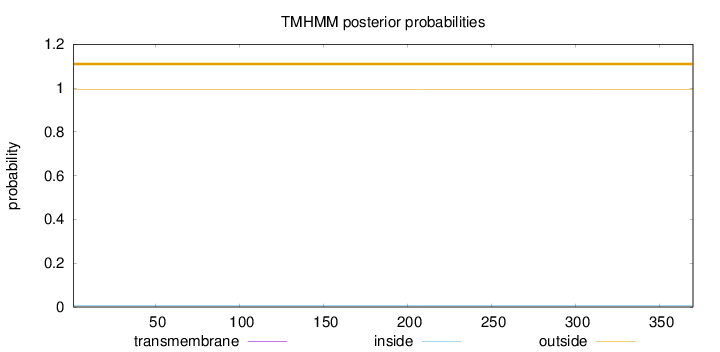
Length:
370
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.003
Exp number, first 60 AAs:
0.00117
Total prob of N-in:
0.00830
outside
1 - 370
Population Genetic Test Statistics
Pi
11.066635
Theta
3.865061
Tajima's D
-1.523534
CLR
16.890825
CSRT
0.0517474126293685
Interpretation
Uncertain
