Gene
KWMTBOMO12667 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001402
Annotation
mitochondrial_aldehyde_dehydrogenase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.277 Mitochondrial Reliability : 1.767 Nuclear Reliability : 1.15
Sequence
CDS
ATGTTCGGCATCAGAACACCAGTTAAAAAACCAAACGGCAGTGAGGACAAGTCTGAATTCGAAGGACAGCCAGGTCCGTCTGTAAGGCGCAGCATTGGTGATTGGCCCCCCATAGTCGAAAAAGAGCAGCAAAGGCCAAAAACACCATCTCCACCAAAAGCTGTGCCTCAGCCGCTCCCTCCTAGGCCTAAGACCAAAGCGTTGATCGCCCAGGAAGCAAAAGCTGACCCACGGAGACAGGCGGCAAGAGTCAGTGACAGCCCGCCAATTACAGTCCCGTCGAGATTTCCGAGCAAAACCGCAGAGGCTAAAGCGTGCCTTATGAAAAGCAAGGCGGAGCTCGAAATCTCCAAAAACCTAAAACGGGAGATAAAAGAAGAAGTCTTTAGGTCAATCGAACGGCTCTACCAGCTGGTGAAAGAAGCCGAAGGCGAAAAGGCTTCCCAAGCCACCAACAAGTGGCCGACAGAAGCCTGTAGCATGGTCACTCGGACTGGTAACGAGGGTGACATATCACCGGGCCCAGAAGCGCCGACAGAGCTCATGAAACAGCTAATAGGCGGCCTCAAACAGCACCACGTTGCGTTAGAGGAATGCAAACGGCATACTCGAGCTCTGTCGGAACAGCTGGAGAAAACCCCTCTCTGCGCCGCAGGACATGGGTCATTAGATGAACACAGCCAACTCTTAAAGGAAAACAAAGAAGAAATAGAAAAACTAAGTGAAGAAATAAAAAAGCAAAGGACAGAATGGAAAGAAAATAGGAGCTACGCAAGCGTTGTCAGTGGCACAAAGGAGAAAGAACGCGGAAATGCGGAGATGCATTCCGTCGTGATCTCCTCAGACGACCAGAATGCCACAGGGGAAGACGTATTTAATAAGATCAGGTCAACCCTCAACGCTAAAGAGGAAGGGATAAAAATAGACAGGATAAGAAAGGTGAAAGATAGAAAAGTTATAGTCGGCTGTGAATCGAAAGAAGAATTAGTAAAAATAAGAGAAACAATTAAAACAAAGGGAAAGGACTTACACCTCCAGGAGATAAGAAATAAGGAACCCATGGTACAATTGAAGGATGTCCTGAATTATAATAAGGATGAAGATATAGTGCGAGCACTAATTAACCAGAACAAACATCTTCTGGGGGATGTGAGTGTGGACAGCTCAACTCTGAAGGTAAAGTTCAGAAGAAGAGCTAGAAACCCGCTCACATCCCATGTGGCAATACAAGTGCCACCAAAACTATGGAAGCGGCTCACGGAGGCGGGGCTAGTCCATATAGATGTGCAAAGGGTCAGGGTAGAGGATCAGTCTCCCCTGATTCAGTGCTCGCGATGCTTGGGATATGGCCACGGGAAGCGGTTCTGCCGGGAAAAAATTGACGCCTGCAGTCACTGCGGAGAGGCGCATCATAGGTCGGAATGCGCTGACTGGATGGCGAGGGTGCCTCCGTCGTGCTGTAACTGCTTGAAGGCTAAAATCGGTGAAGCCGAGCATAATGCCTTTAGCCCAGAGTGTCCAGTAAGGAAGAGACTTGGATCACCATGGAGAACAATGGACGCCTCCGAGAGGGGGGCCCTTATCAACAAACTAGCAGATTTGATTGAAAGAGACAGAACCTATTTAGCTAGCCTGGAGACGCTGGACAACGGCAAACCATACAAGGACTCGTATTTCGGTGATCTGTATGCTTCAATAAAAAACCTACGGTACTACGCCGGGTGGGCCGACAAGATCCACGGGAATGTCCTGCCCGCCGACGGAAAATACTTTGCTTACACTCGTCATGAGCCAGTGGGTGTGTGCGGTCAGATAATACCATGGAACTTCCCTATATTGATGGCGGCTTGGAAGCTCGGGCCAGCTCTGGCGACGGGTTGTACTGTGGTCATGAAGCCGGCCGAGCAGACGCCGCTCACAGCTCTGTACATCGCTCAACTAGTCAAGGAAGCAGGTTTCCCACCCGGGGTCGTGAATATGTTGCCGGGCTACGGAGACACCGGGGCCGCCATCGTGGACCACCCCGACGTAGATAAAGTGGCCTTCACCGGTTCGACGGAGGTGGGCAAGCTAATCCAGCGCGGCGCGGCGTCGACGATAAAGCGCATCACGCTGGAGCTGGGCGGCAAGTCGCCCAACATCGTGCTGGCCGACACCGACCTGCCGCGCGCCGTGGAGGCCGCGCACAACGCGCTCTTCTACAACATGGGGCAGTGCTGCTGCGCCGGCTCTCGCACCTTCGTGGAGGACAAGATTTACGACCAGTTCGTAGAGTTGTCGGCCGAGCGCGCCAACCGCCGCGTGGTCGGGGACCCCTTCCGGCCGGACGTGGAGCAGGGACCCCAGATCGACAACGAGCAGCGCGACAAGATCCTGCAGCTGATCAGCTCCGGGTCGCGGCAGGGCGCGCGGCTGGCGGCGGGCGGCGGCGCGGCCCCCGGGCCCGGCTACTTCGTGCAGCCCACCGTCTTCTGTGACGTCACCGACGACATGGACATCGCTAAGACAGAGATCTTTGGGCCGGTGCAGCAAATCCTGAAGTTCTCCCAGTTCGACGAGGTGGTGGAGCGCGCCAACAACTCGGAGTACGGACTGGCCGCCGCTGTCTTCACTAAAGATCTAGACAAGGCCAACTACTTCGTACAGAGGCTGCGCGCCGGCACCATTTGGGTTAACGACTACAACGTATTCGGGAACCAAGTGCCCTTCGGCGGCTTCAAGCAGTCCGGGCTGGGCCGCGAGAACGGGCCCTACGGACTCCGCAACTACCTCGAGGTCAAGGCCGTCGTCGTCAAACTCGCCGACAAGAACACGTGA
Protein
MFGIRTPVKKPNGSEDKSEFEGQPGPSVRRSIGDWPPIVEKEQQRPKTPSPPKAVPQPLPPRPKTKALIAQEAKADPRRQAARVSDSPPITVPSRFPSKTAEAKACLMKSKAELEISKNLKREIKEEVFRSIERLYQLVKEAEGEKASQATNKWPTEACSMVTRTGNEGDISPGPEAPTELMKQLIGGLKQHHVALEECKRHTRALSEQLEKTPLCAAGHGSLDEHSQLLKENKEEIEKLSEEIKKQRTEWKENRSYASVVSGTKEKERGNAEMHSVVISSDDQNATGEDVFNKIRSTLNAKEEGIKIDRIRKVKDRKVIVGCESKEELVKIRETIKTKGKDLHLQEIRNKEPMVQLKDVLNYNKDEDIVRALINQNKHLLGDVSVDSSTLKVKFRRRARNPLTSHVAIQVPPKLWKRLTEAGLVHIDVQRVRVEDQSPLIQCSRCLGYGHGKRFCREKIDACSHCGEAHHRSECADWMARVPPSCCNCLKAKIGEAEHNAFSPECPVRKRLGSPWRTMDASERGALINKLADLIERDRTYLASLETLDNGKPYKDSYFGDLYASIKNLRYYAGWADKIHGNVLPADGKYFAYTRHEPVGVCGQIIPWNFPILMAAWKLGPALATGCTVVMKPAEQTPLTALYIAQLVKEAGFPPGVVNMLPGYGDTGAAIVDHPDVDKVAFTGSTEVGKLIQRGAASTIKRITLELGGKSPNIVLADTDLPRAVEAAHNALFYNMGQCCCAGSRTFVEDKIYDQFVELSAERANRRVVGDPFRPDVEQGPQIDNEQRDKILQLISSGSRQGARLAAGGGAAPGPGYFVQPTVFCDVTDDMDIAKTEIFGPVQQILKFSQFDEVVERANNSEYGLAAAVFTKDLDKANYFVQRLRAGTIWVNDYNVFGNQVPFGGFKQSGLGRENGPYGLRNYLEVKAVVVKLADKNT
Summary
Uniprot
ProteinModelPortal
PDB
4KWG
E-value=8.46138e-171,
Score=1545
Ontologies
GO
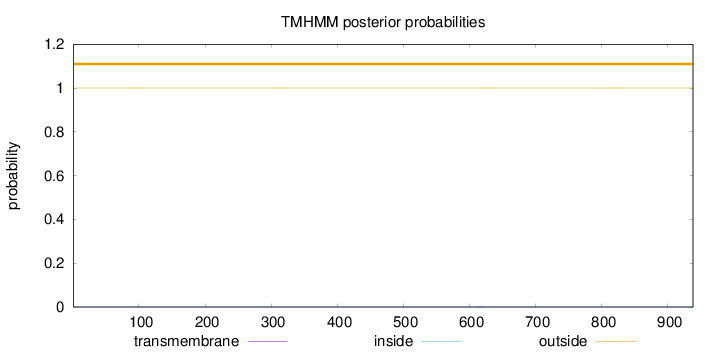
Topology
Length:
938
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0054
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00003
outside
1 - 938
Population Genetic Test Statistics
Pi
12.787476
Theta
17.267288
Tajima's D
-1.537615
CLR
131.129992
CSRT
0.056197190140493
Interpretation
Uncertain
