Gene
KWMTBOMO12666
Pre Gene Modal
BGIBMGA001577
Annotation
PREDICTED:_uncharacterized_protein_LOC106129070_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.226
Sequence
CDS
ATGGGAGTATTCAAGTGGTTACGTCGGGAACGTGTGACTGTGACCACTCGTCACAGCGTCAGCCCGACGTCCACAGTTCCAGAGGAACAGAATGCACGAGACCCGCAAATAAACGCCGTCCGCACTGAAGAGTACAAACGTGATTTAGCGGAGTTCCAAGCCAGACGTTCTATTGGGAACAGAGATCTACAAGAAACTCCGGTTAAAGAAAAACGTTCACGACACTCTCTTCCGGTTTCGTGCACCGACTGTTCCGCAGATCACGATAACACCTTCAACTATGAAAAAAAATCCGCTATCAAATACAAAAAGAAACAAGATCAGCGCAATCGAAGTACAACACCAGAGAAAGTGTACGAAGGCGTTCCGAACATACAATTTTTGTTCCAGAACCAAGTGTTCATGCCCGGGAATATGTTTCCGAACCCTTCGTATTCTGAACCCCATAAGACCCACCACCGCACCCGACGACACGTCTGTCGATCTCCCGATTTGGATTTCGTTGCTCAGAAACACATTGACTTCCACGAGGATGATCAGGTTGATTCGCCACCTGATTTCTACAAGAATAGTTCCTTGCCTTTTGAAACGAAAGACTTTCTGGGAACTTCCAGAATATTCCACGAAGAGGTGGACGGCTTGAATAAAGCGGGCGACGTGAAATCCTTCAATAATTCGGAATCCGAAAGTGTCAACAAAGATGACAGCTGTGAAGATAAGCTATGGGAGGTAATGAGCGAGTTGAAGAACTTTGATCAATGGGCGGATGAGCAGTTGGTTAGGTCACCAAACACGACCAAATCAGATGACAGCAAGAGCGATACGAGCGCTTTTGGTTTGCCAATTGCGAGTGCCAGGAGTCTCGACATGACAATAGATAAGGCTAAGACGTGGAATAACGGCAAGTGGGGTGTAGTTCCGGTTCAGATTAAGAAGTTGGTCGGTGTAACCGCTCAACATGTTAGGAAGAAACGGAACACCGAGATTAATATATTACGAAAATGTCGTCACCCGAACATTATCTTATTGATGGGCCTGTATCCGGATGCACAGAACAACATTCACTTAATCTGCGAACGATGCGTCGATACCTTATATGGAATTATTCACGAAAAGGGTCGCATACTAAGCGCACAGACGGCGGTGCAGTACGCACTGGACATATCGAATGCTCTAGTTTTCTTGCGAATGCAAGGTTTCATACACACGGAACTGAACAGCGCGTGTGTAATGATAACGGATCACGACACGGCCAAGCTGGTCGACGTGGGTCCCTGTACGAAGGCGCCGAGAACATTACTCAAACGAGAGAAAGAATATGAGGTATACAACCCCGAACCGCAGTATGAGGCCATCCAAGATAGTGCCCGTGTAAAACCGAATGACTGTGAATCTCTCATCTCCGAAAATCAATCTGAATTATACAGATTATCGAACTGCGTTGAAAAATCATATAAATCGTATGAGACTTCGGAATTTTACAACTGGCACGCTCCGGAATTGTTTACACCTAACGACGCAGGGGTCGTGTACGTGTGCTCCAGAAGCGACGTTTATTCGCTCTGTTTGATTCTCTGGGAATGTTGTAACGCATTACGACCTTGGAAGGACTTTAGCCGCGAAAAGCTGAAGGAACAGTACACGTTGTGGAAGTCCGGCGTTAACTTACCTAATGATGGAACTTATCCGGGTTGTCTGCTGGCGCTCCTGCAGAGTGGCTTGAAAGTACAAAGGAACGAACGCATCGACTTGGACACATTGCAAAAAACTTTGCAAAACGTAAAGCGCGACCTGGATGACTTGGGTTACTTTATAATACCGGCACGGCTTCCTCACAAAAAGGCTACATTCAGCACTGCCGAATGGGAAACAGCATCACCTGCGGCTTCTGAATGTCAGAGTCCAAAAAACGTAGAACATAGAGCAGTTGTTCACGACCGCCATGACAGTTCCACTGAGAGCAGCGAGAAAGAATGCACTCGAAGCGACGCCAGCCCCGTTTACGAAACCATCAAAAAAGATTACGAAGACCCAATATACTCGTACCCGGAAAAAAGTTTCGTAAACGACAAGAAGATGTCTTTACAATACGATCGTTTGTCGGAAAAGGGATTGTCTAGTGCGTGCTCGACACCGATCACTAAAATACGAGCAAGAATTAAAGACATCGGGACCCCCGGAACTCTGCATCGGAGCGATTCCACAGAGTACTGTAGCATATTCAGTCCGAATCGAACGGTCAATTTTGAACTCAACAACAATATAAATAGAGATCAAAGCGTCGAGCGCCCTCTCTCTAAGCTAAGTAAGAGCTTCACGCCACAGTCGTACAAGCCCTTACACATAAAAGTTCCAGAGTACAATTTCGATTCTATACGTCCTATTTTGAAAGAGCCCACCGGCTCTGAGCGCCCGTCGTACAATTTCGATATAAAAAACTACAGTTTGCCGACGACGCCTATAGCAAGGAGTACGAAGTTGAGAAGAAATGCTTGGCTTTCTGGGGACTTAAATATTTCCGATCGATGTTTCGAGAAATCAAAAGAGAACCTCTTAAAGTCAATGGATAAAGAAGTTCGCTCAGCACCTGAAACTCCAGCCACGTCTGTATGTTCAATGTCGGAGAAACCATATTATTCTGAAAAAAGTAGTCCCGAAGAAGATAAAAATGAAAGGACTCCTGAGAGCGTGGATACCATTCGAGATTGCTTTACGGGACTTAAACGAATCGAACATCCACACGACTTTGTGTCGTCGGAGACGGACGAGACGTGTAGAGAACGAGCAAATTCATGGTCATTCAAATCGTCTTCCGAACGCGGCCCGAAATACAAAATAGAGGATGATTTATTGGCGAGAGTCAACGTCAAGCCGCTCGTCGCGATACACGAGCGATGGATATACGAGGCCAATAGGAAGAACAGCCGTTCGATGTCCTTACCAGAAGATAAAAGGTGTCAAATTCCGGCCGTCAAACCGGCGTCCCACTGCGTAGACACTCTGCAGAAGTCTCTGCCACAGCTGAATCTGAAGAAGTCTGGTACTAAAATATATAGGTCTTGCGCTGCCTCTCCGAATAGTAATACTAAAAACGATGTTCAGTGGGAGCAATTCTGTCAAGCGCGGGACGCGATATCCAAGAATGTTTCTTTCCTGCAGAAGGAATCTGATCGATCACTTGTACAAAGACGGGATTCTAGTAATACATCCATAGAAGAGAAATATGAAAGAAAGATAGACCAGGCGACGGACACGAACGGCATCGAGGATTTTATAAGGGACATGATCCGCAAGGAATTCAAACACTTGCTCCACGAACTATCCGACGACGCGTCCACTACGTCAGCGAACCGACACGAAGACATCCTCACCAAGGGCGTCATTGAATCTCTGAACGGCGCCGAAGACGACAAAACCACAAAAAGGGAACCGATCAAATCGTTTTCGCGAGTCAAAATCAACGGGAACAATAAACCGCTTCTACAGATCACCTTCAGCCGCTCGGGCGAGAACAGTCTGCAGGTCCTGGACAACTGCGTCAACGTGACCGTCTGCGACACCGGCTCGCCCAGGGACGCGCCCGCCGACGACGTGCACCTCAACTCACTGAAGAGGTGTCAGGCTTTCAACGCTATTCAGGAAGCAAGAAGTACAGAAGATTTGTACATAGACGACGATCTATCGACGCAGATGCAGGAGAACTTCGGCGGACGGGTCAAGCTGATCCCCCTGCACGGGTCCTTGCACGAGATCCTGTGCGAGAAGGAAGACGACTGCTGCCTGATCAAAGTGAAGCAGGAAAACGGATGCGACTCCATATACTTCCGCTGCGACAATTCCGACACCGAGTCCAGGGACGCCATTGACGGGACGCAAGACACGGTCGTCTTCAAGCGGAGCATCTCTCTCATAGAAGAACGAATACAGAGGGTGCCCAAAGAGAAACGGGCCCCCACGCCGATACTGCAAAGGAAAGCTAAACCGGACAAGGCACGACCCAAATCAGAAATATTTATTCCCATCAAAAGGGAGTCTCGCGCCATGAGCAACAGCAGTCCCTCGCTGTGCGCGCGAGATGACAAGGACGAGGTTGAGGTCAACATCGAGAACGCGATGTGCTACCAGGACTCTTCGTCCGACGAGTGTCTCAAATGTCAGGACAATGACGACTTCGAATTTCTCGAGCGAAAAATCGAAGAGGACATCGCCAACAACCTGTCTTCGCTCGCTTGCGGGTGCCTAGAGAAAATATCCGAAGAGAATCTCTCCGTTTTTAACGAAGATTTGGGTGATAAAGAGTGA
Protein
MGVFKWLRRERVTVTTRHSVSPTSTVPEEQNARDPQINAVRTEEYKRDLAEFQARRSIGNRDLQETPVKEKRSRHSLPVSCTDCSADHDNTFNYEKKSAIKYKKKQDQRNRSTTPEKVYEGVPNIQFLFQNQVFMPGNMFPNPSYSEPHKTHHRTRRHVCRSPDLDFVAQKHIDFHEDDQVDSPPDFYKNSSLPFETKDFLGTSRIFHEEVDGLNKAGDVKSFNNSESESVNKDDSCEDKLWEVMSELKNFDQWADEQLVRSPNTTKSDDSKSDTSAFGLPIASARSLDMTIDKAKTWNNGKWGVVPVQIKKLVGVTAQHVRKKRNTEINILRKCRHPNIILLMGLYPDAQNNIHLICERCVDTLYGIIHEKGRILSAQTAVQYALDISNALVFLRMQGFIHTELNSACVMITDHDTAKLVDVGPCTKAPRTLLKREKEYEVYNPEPQYEAIQDSARVKPNDCESLISENQSELYRLSNCVEKSYKSYETSEFYNWHAPELFTPNDAGVVYVCSRSDVYSLCLILWECCNALRPWKDFSREKLKEQYTLWKSGVNLPNDGTYPGCLLALLQSGLKVQRNERIDLDTLQKTLQNVKRDLDDLGYFIIPARLPHKKATFSTAEWETASPAASECQSPKNVEHRAVVHDRHDSSTESSEKECTRSDASPVYETIKKDYEDPIYSYPEKSFVNDKKMSLQYDRLSEKGLSSACSTPITKIRARIKDIGTPGTLHRSDSTEYCSIFSPNRTVNFELNNNINRDQSVERPLSKLSKSFTPQSYKPLHIKVPEYNFDSIRPILKEPTGSERPSYNFDIKNYSLPTTPIARSTKLRRNAWLSGDLNISDRCFEKSKENLLKSMDKEVRSAPETPATSVCSMSEKPYYSEKSSPEEDKNERTPESVDTIRDCFTGLKRIEHPHDFVSSETDETCRERANSWSFKSSSERGPKYKIEDDLLARVNVKPLVAIHERWIYEANRKNSRSMSLPEDKRCQIPAVKPASHCVDTLQKSLPQLNLKKSGTKIYRSCAASPNSNTKNDVQWEQFCQARDAISKNVSFLQKESDRSLVQRRDSSNTSIEEKYERKIDQATDTNGIEDFIRDMIRKEFKHLLHELSDDASTTSANRHEDILTKGVIESLNGAEDDKTTKREPIKSFSRVKINGNNKPLLQITFSRSGENSLQVLDNCVNVTVCDTGSPRDAPADDVHLNSLKRCQAFNAIQEARSTEDLYIDDDLSTQMQENFGGRVKLIPLHGSLHEILCEKEDDCCLIKVKQENGCDSIYFRCDNSDTESRDAIDGTQDTVVFKRSISLIEERIQRVPKEKRAPTPILQRKAKPDKARPKSEIFIPIKRESRAMSNSSPSLCARDDKDEVEVNIENAMCYQDSSSDECLKCQDNDDFEFLERKIEEDIANNLSSLACGCLEKISEENLSVFNEDLGDKE
Summary
Uniprot
Pubmed
Proteomes
Pfam
PF07714 Pkinase_Tyr
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
PDB
4XV3
E-value=4.89059e-07,
Score=134
Ontologies
GO
PANTHER
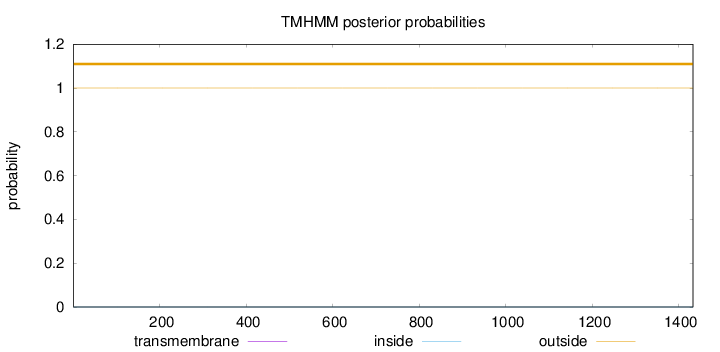
Topology
Length:
1433
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000610000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00004
outside
1 - 1433
Population Genetic Test Statistics
Pi
20.283881
Theta
21.955542
Tajima's D
-0.83163
CLR
0.987176
CSRT
0.169191540422979
Interpretation
Uncertain
