Gene
KWMTBOMO12664
Pre Gene Modal
BGIBMGA001403
Annotation
PREDICTED:_uncharacterized_protein_LOC106129089_isoform_X2_[Amyelois_transitella]
Full name
Centrosomal protein of 120 kDa
Alternative Name
Coiled-coil domain-containing protein 100
Location in the cell
Cytoplasmic Reliability : 1.887 Nuclear Reliability : 2.268
Sequence
CDS
ATGTACTTACTGGAGCCCGTCGTTTTGGAGACGAGCAAAAGCGACACGTGCATCACAAGTCCTAAACGGGCCGCCCGTAAAGACAATCACACAGATTTGGTGAAAATGGCGCTATCGGAAGCCGACCGCGACAGCATCATGCGGAGGTTCGTGGACGAGCTGGAAGACTGGAAGGAGAAGCAGCAGGAGCTGCACAAACTGCAGTTGAAGCGCAAAGAAGAGTACCATTTAGAGCTATTGGCCAAGGAGTGGGCCAAGCAGCGCGTGGAGCTGGAGTGCCGGCTGGCGAGGGGCATCGAGCAGTGCCGCGCCCTCGCCGCCGACCTCGCCGCCGCCACAGACGACTTCAGGGCGAGGGGACACAGGAACTCCGAGAGGGAGAAGAAGTTATTGGAAGCGAAGAAAGCTTTAGAAGCGCATTACACAGCCAAGTACCAGGAGTTGAGAGAGGCCTCATTAAAAATGGAGGACGACATGAATCATCAGCTGAAAGTCAAGGACATGAGGATCGAAGAGCTGGAGCTTAAAATAATGCAGCTTGAGAAACAGGTGGACGTTTTGAAGAATACCATGAAGGACATTGAGAAAGAAGCCGAAACCCGATATTCTGGTCTAACTAAAGATCAAACTGCGAGTCTCATACAAGAATTGAGATGTTTAGAAGAAAAACTAGATAGTGCGGTCCAATCGAAAGCGTTCTTCAAAGAGCAGTGGGGCCGCGCTGTCAGGGAACTGCACCTGCTGAAGCTGGACACCAGGAAGCAAATGTTGTCGCAGCTGCAGCAGGACAGGAGGGAGCTGGGTGATGCTGGTTTAGACACAATAATAGATGACAACGAAAGTAAACGCAATCAAGATGCGATGGACATTAAAAAATTGAAAGACGATTTTTATGTTGACATTTTAGCGAGCACACCGGCACTGGAAATCGATTCGTTCATCACCACATCTGGTTCGGGTGTTGTAGAAATGTTTGACGATTTAAGAAGCGTCGTTAAGAACCCGATAAATGATAAACTAAACAAATTGATATCACAACGCGATCGGCTCATACAGGACGAGAATCCGAATGAAGAACTTCTGAAGCAATTGAACCAGGAAATTAGAAGTATCCTATTAAACTGTGGCTCCTGA
Protein
MYLLEPVVLETSKSDTCITSPKRAARKDNHTDLVKMALSEADRDSIMRRFVDELEDWKEKQQELHKLQLKRKEEYHLELLAKEWAKQRVELECRLARGIEQCRALAADLAAATDDFRARGHRNSEREKKLLEAKKALEAHYTAKYQELREASLKMEDDMNHQLKVKDMRIEELELKIMQLEKQVDVLKNTMKDIEKEAETRYSGLTKDQTASLIQELRCLEEKLDSAVQSKAFFKEQWGRAVRELHLLKLDTRKQMLSQLQQDRRELGDAGLDTIIDDNESKRNQDAMDIKKLKDDFYVDILASTPALEIDSFITTSGSGVVEMFDDLRSVVKNPINDKLNKLISQRDRLIQDENPNEELLKQLNQEIRSILLNCGS
Summary
Description
Plays a role in the microtubule-dependent coupling of the nucleus and the centrosome. Involved in the processes that regulate centrosome-mediated interkinetic nuclear migration (INM) of neural progenitors and for proper positioning of neurons during brain development. Also implicated in the migration and selfrenewal of neural progenitors. Required for centriole duplication and maturation during mitosis and subsequent ciliogenesis. Required for the recruitment of CEP295 to the proximal end of new-born centrioles at the centriolar microtubule wall during early S phase in a PLK4-dependent manner (By similarity).
Plays a role in the microtubule-dependent coupling of the nucleus and the centrosome. Involved in the processes that regulate centrosome-mediated interkinetic nuclear migration (INM) of neural progenitors and for proper positioning of neurons during brain development. Also implicated in the migration and selfrenewal of neural progenitors. Required for centriole duplication and maturation during mitosis and subsequent ciliogenesis (By similarity). Required for the recruitment of CEP295 to the proximal end of new-born centrioles at the centriolar microtubule wall during early S phase in a PLK4-dependent manner (PubMed:27185865).
Plays a role in the microtubule-dependent coupling of the nucleus and the centrosome. Involved in the processes that regulate centrosome-mediated interkinetic nuclear migration (INM) of neural progenitors and for proper positioning of neurons during brain development. Also implicated in the migration and selfrenewal of neural progenitors. May play a role in centriole duplication during mitosis (By similarity). Required for the recruitment of CEP295 to the proximal end of new-born centrioles at the centriolar microtubule wall during early S phase in a PLK4-dependent manner (By similarity).
Plays a role in the microtubule-dependent coupling of the nucleus and the centrosome. Involved in the processes that regulate centrosome-mediated interkinetic nuclear migration (INM) of neural progenitors and for proper positioning of neurons during brain development. Also implicated in the migration and selfrenewal of neural progenitors. Required for centriole duplication and maturation during mitosis and subsequent ciliogenesis (By similarity). Required for the recruitment of CEP295 to the proximal end of new-born centrioles at the centriolar microtubule wall during early S phase in a PLK4-dependent manner (PubMed:27185865).
Plays a role in the microtubule-dependent coupling of the nucleus and the centrosome. Involved in the processes that regulate centrosome-mediated interkinetic nuclear migration (INM) of neural progenitors and for proper positioning of neurons during brain development. Also implicated in the migration and selfrenewal of neural progenitors. May play a role in centriole duplication during mitosis (By similarity). Required for the recruitment of CEP295 to the proximal end of new-born centrioles at the centriolar microtubule wall during early S phase in a PLK4-dependent manner (By similarity).
Subunit
Interacts with TACC2, TACC3, CCDC52, TALPID3.
Interacts with TACC2 and TACC3. Interacts with CCDC52.
Interacts with TACC2 and TACC3. Interacts with CCDC52.
Similarity
Belongs to the CEP120 family.
Keywords
3D-structure
Alternative splicing
Coiled coil
Complete proteome
Cytoplasm
Cytoskeleton
Phosphoprotein
Reference proteome
Ciliopathy
Disease mutation
Joubert syndrome
Polymorphism
Feature
chain Centrosomal protein of 120 kDa
splice variant In isoform 4.
sequence variant In JBTS31; unknown pathological significance.
splice variant In isoform 4.
sequence variant In JBTS31; unknown pathological significance.
Uniprot
H9IVX2
A0A2A4IW85
A0A2H1VQW2
A0A3S2NLW6
A0A194RD83
A0A212FFR2
+ More
A0A0J7KNJ4 E2A1Y5 A0A158NBU0 A0A0N0BDS7 F4X898 A0A195C9L5 A0A151J942 A0A0C9QLD1 A0A0C9PK77 A0A232FAE4 K7IP62 A0A026W5Q7 A0A0L7QLA1 A0A3L8DWD3 A0A2A3ERL2 A0A088AUH2 A0A151X9N6 E2BIF1 A0A067QR70 A0A195BF06 E0VRM6 A0A1B6CTJ9 K1QC39 A0A1B6DNS3 A0A1S3HGB8 A0A2K6UN37 A0A2K6UMX9 A0A2K5RRD1 Q7TSG1 A0A2K6CKX9 L5LUF1 A0A2T7NEV3 A0A2K6A166 A0A2J8LKA2 Q8N960-2 A0A2K6A1B3 H2QRE4 G3QZS3 K7CF68 Q8N960 A0A2K5K9C3 G3T5U6 A0A2J8XE72 A0A2J8XE97 A0A2K5K9C6 A0A0D9RQ03 A0A2K5LAN3 A0A452E7G0 W5QBW0 A0JN62 A0A0P6JF49 G5C983 A0A2R9ASC9 D4A3J3 U3D6X8 F7I5B6 A0A0B8RT39 F1RKN9
A0A0J7KNJ4 E2A1Y5 A0A158NBU0 A0A0N0BDS7 F4X898 A0A195C9L5 A0A151J942 A0A0C9QLD1 A0A0C9PK77 A0A232FAE4 K7IP62 A0A026W5Q7 A0A0L7QLA1 A0A3L8DWD3 A0A2A3ERL2 A0A088AUH2 A0A151X9N6 E2BIF1 A0A067QR70 A0A195BF06 E0VRM6 A0A1B6CTJ9 K1QC39 A0A1B6DNS3 A0A1S3HGB8 A0A2K6UN37 A0A2K6UMX9 A0A2K5RRD1 Q7TSG1 A0A2K6CKX9 L5LUF1 A0A2T7NEV3 A0A2K6A166 A0A2J8LKA2 Q8N960-2 A0A2K6A1B3 H2QRE4 G3QZS3 K7CF68 Q8N960 A0A2K5K9C3 G3T5U6 A0A2J8XE72 A0A2J8XE97 A0A2K5K9C6 A0A0D9RQ03 A0A2K5LAN3 A0A452E7G0 W5QBW0 A0JN62 A0A0P6JF49 G5C983 A0A2R9ASC9 D4A3J3 U3D6X8 F7I5B6 A0A0B8RT39 F1RKN9
Pubmed
19121390
26354079
22118469
20798317
21347285
21719571
+ More
28648823 20075255 24508170 30249741 24845553 20566863 22992520 16141072 15489334 17920017 21183079 20360068 25251415 15372022 14702039 17974005 14654843 23186163 24275569 25361962 27185865 27208211 16136131 22398555 20809919 19390049 21993625 22722832 15057822 22673903 25243066 30723633
28648823 20075255 24508170 30249741 24845553 20566863 22992520 16141072 15489334 17920017 21183079 20360068 25251415 15372022 14702039 17974005 14654843 23186163 24275569 25361962 27185865 27208211 16136131 22398555 20809919 19390049 21993625 22722832 15057822 22673903 25243066 30723633
EMBL
BABH01006497
BABH01006498
NWSH01006503
PCG63390.1
ODYU01003897
SOQ43231.1
+ More
RSAL01000006 RVE54266.1 KQ460325 KPJ15798.1 AGBW02008777 OWR52569.1 LBMM01005028 KMQ91887.1 GL435863 EFN72551.1 ADTU01011323 KQ435850 KOX70862.1 GL888932 EGI57172.1 KQ978068 KYM97554.1 KQ979433 KYN21585.1 GBYB01001427 JAG71194.1 GBYB01001428 JAG71195.1 NNAY01000633 OXU27297.1 KK107459 EZA50369.1 KQ414929 KOC59380.1 QOIP01000004 RLU24249.1 KZ288192 PBC34330.1 KQ982373 KYQ57030.1 GL448504 EFN84539.1 KK853055 KDR11964.1 KQ976500 KYM83171.1 DS235478 EEB16032.1 GEDC01020745 JAS16553.1 JH817066 EKC26375.1 GEDC01010015 JAS27283.1 AK038916 AK044808 AK044952 AK047594 AK079319 BC046493 BC048590 BC053439 KB107782 ELK29721.1 PZQS01000013 PVD19694.1 NBAG03000288 PNI47689.1 AC010369 AC106792 BC040527 AK093409 AK095646 AL833929 BX648687 CR627324 AACZ04009925 GABC01006869 GABD01006534 GABE01010287 JAA04469.1 JAA26566.1 JAA34452.1 PNI47687.1 PNI47688.1 CABD030040669 CABD030040670 GABF01004588 JAA17557.1 NDHI03003367 PNJ80334.1 ABGA01000616 ABGA01000617 ABGA01000618 ABGA01000619 ABGA01000620 PNJ80332.1 PNJ80339.1 AQIB01044001 LWLT01000008 AMGL01092062 AMGL01092063 AMGL01092064 AMGL01092065 AMGL01092066 AMGL01092067 AMGL01092068 AMGL01092069 AMGL01092070 AMGL01092071 AAFC03099167 AAFC03125893 AAFC03125142 AAFC03099169 AAFC03131164 AAFC03126660 BC126531 GEBF01004354 JAN99278.1 JH174046 EHB18094.1 AJFE02093464 AABR07032133 GAMR01007903 JAB26029.1 GAMT01004054 GAMS01010370 GAMP01007073 JAB07807.1 JAB12766.1 JAB45682.1 GBZA01000124 JAG69644.1 AEMK02000012 DQIR01096726 DQIR01163681 DQIR01253284 DQIR01258638 DQIR01258639 DQIR01269033 DQIR01269034 DQIR01269035 DQIR01277159 DQIR01281463 DQIR01285038 DQIR01287666 HDA52202.1 HDB19158.1
RSAL01000006 RVE54266.1 KQ460325 KPJ15798.1 AGBW02008777 OWR52569.1 LBMM01005028 KMQ91887.1 GL435863 EFN72551.1 ADTU01011323 KQ435850 KOX70862.1 GL888932 EGI57172.1 KQ978068 KYM97554.1 KQ979433 KYN21585.1 GBYB01001427 JAG71194.1 GBYB01001428 JAG71195.1 NNAY01000633 OXU27297.1 KK107459 EZA50369.1 KQ414929 KOC59380.1 QOIP01000004 RLU24249.1 KZ288192 PBC34330.1 KQ982373 KYQ57030.1 GL448504 EFN84539.1 KK853055 KDR11964.1 KQ976500 KYM83171.1 DS235478 EEB16032.1 GEDC01020745 JAS16553.1 JH817066 EKC26375.1 GEDC01010015 JAS27283.1 AK038916 AK044808 AK044952 AK047594 AK079319 BC046493 BC048590 BC053439 KB107782 ELK29721.1 PZQS01000013 PVD19694.1 NBAG03000288 PNI47689.1 AC010369 AC106792 BC040527 AK093409 AK095646 AL833929 BX648687 CR627324 AACZ04009925 GABC01006869 GABD01006534 GABE01010287 JAA04469.1 JAA26566.1 JAA34452.1 PNI47687.1 PNI47688.1 CABD030040669 CABD030040670 GABF01004588 JAA17557.1 NDHI03003367 PNJ80334.1 ABGA01000616 ABGA01000617 ABGA01000618 ABGA01000619 ABGA01000620 PNJ80332.1 PNJ80339.1 AQIB01044001 LWLT01000008 AMGL01092062 AMGL01092063 AMGL01092064 AMGL01092065 AMGL01092066 AMGL01092067 AMGL01092068 AMGL01092069 AMGL01092070 AMGL01092071 AAFC03099167 AAFC03125893 AAFC03125142 AAFC03099169 AAFC03131164 AAFC03126660 BC126531 GEBF01004354 JAN99278.1 JH174046 EHB18094.1 AJFE02093464 AABR07032133 GAMR01007903 JAB26029.1 GAMT01004054 GAMS01010370 GAMP01007073 JAB07807.1 JAB12766.1 JAB45682.1 GBZA01000124 JAG69644.1 AEMK02000012 DQIR01096726 DQIR01163681 DQIR01253284 DQIR01258638 DQIR01258639 DQIR01269033 DQIR01269034 DQIR01269035 DQIR01277159 DQIR01281463 DQIR01285038 DQIR01287666 HDA52202.1 HDB19158.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000036403
+ More
UP000000311 UP000005205 UP000053105 UP000007755 UP000078542 UP000078492 UP000215335 UP000002358 UP000053097 UP000053825 UP000279307 UP000242457 UP000005203 UP000075809 UP000008237 UP000027135 UP000078540 UP000009046 UP000005408 UP000085678 UP000233220 UP000233040 UP000000589 UP000233120 UP000245119 UP000233140 UP000005640 UP000002277 UP000001519 UP000233080 UP000007646 UP000001595 UP000029965 UP000233060 UP000291000 UP000002356 UP000009136 UP000006813 UP000240080 UP000002494 UP000008225 UP000008227
UP000000311 UP000005205 UP000053105 UP000007755 UP000078542 UP000078492 UP000215335 UP000002358 UP000053097 UP000053825 UP000279307 UP000242457 UP000005203 UP000075809 UP000008237 UP000027135 UP000078540 UP000009046 UP000005408 UP000085678 UP000233220 UP000233040 UP000000589 UP000233120 UP000245119 UP000233140 UP000005640 UP000002277 UP000001519 UP000233080 UP000007646 UP000001595 UP000029965 UP000233060 UP000291000 UP000002356 UP000009136 UP000006813 UP000240080 UP000002494 UP000008225 UP000008227
Gene 3D
ProteinModelPortal
H9IVX2
A0A2A4IW85
A0A2H1VQW2
A0A3S2NLW6
A0A194RD83
A0A212FFR2
+ More
A0A0J7KNJ4 E2A1Y5 A0A158NBU0 A0A0N0BDS7 F4X898 A0A195C9L5 A0A151J942 A0A0C9QLD1 A0A0C9PK77 A0A232FAE4 K7IP62 A0A026W5Q7 A0A0L7QLA1 A0A3L8DWD3 A0A2A3ERL2 A0A088AUH2 A0A151X9N6 E2BIF1 A0A067QR70 A0A195BF06 E0VRM6 A0A1B6CTJ9 K1QC39 A0A1B6DNS3 A0A1S3HGB8 A0A2K6UN37 A0A2K6UMX9 A0A2K5RRD1 Q7TSG1 A0A2K6CKX9 L5LUF1 A0A2T7NEV3 A0A2K6A166 A0A2J8LKA2 Q8N960-2 A0A2K6A1B3 H2QRE4 G3QZS3 K7CF68 Q8N960 A0A2K5K9C3 G3T5U6 A0A2J8XE72 A0A2J8XE97 A0A2K5K9C6 A0A0D9RQ03 A0A2K5LAN3 A0A452E7G0 W5QBW0 A0JN62 A0A0P6JF49 G5C983 A0A2R9ASC9 D4A3J3 U3D6X8 F7I5B6 A0A0B8RT39 F1RKN9
A0A0J7KNJ4 E2A1Y5 A0A158NBU0 A0A0N0BDS7 F4X898 A0A195C9L5 A0A151J942 A0A0C9QLD1 A0A0C9PK77 A0A232FAE4 K7IP62 A0A026W5Q7 A0A0L7QLA1 A0A3L8DWD3 A0A2A3ERL2 A0A088AUH2 A0A151X9N6 E2BIF1 A0A067QR70 A0A195BF06 E0VRM6 A0A1B6CTJ9 K1QC39 A0A1B6DNS3 A0A1S3HGB8 A0A2K6UN37 A0A2K6UMX9 A0A2K5RRD1 Q7TSG1 A0A2K6CKX9 L5LUF1 A0A2T7NEV3 A0A2K6A166 A0A2J8LKA2 Q8N960-2 A0A2K6A1B3 H2QRE4 G3QZS3 K7CF68 Q8N960 A0A2K5K9C3 G3T5U6 A0A2J8XE72 A0A2J8XE97 A0A2K5K9C6 A0A0D9RQ03 A0A2K5LAN3 A0A452E7G0 W5QBW0 A0JN62 A0A0P6JF49 G5C983 A0A2R9ASC9 D4A3J3 U3D6X8 F7I5B6 A0A0B8RT39 F1RKN9
Ontologies
GO
PANTHER
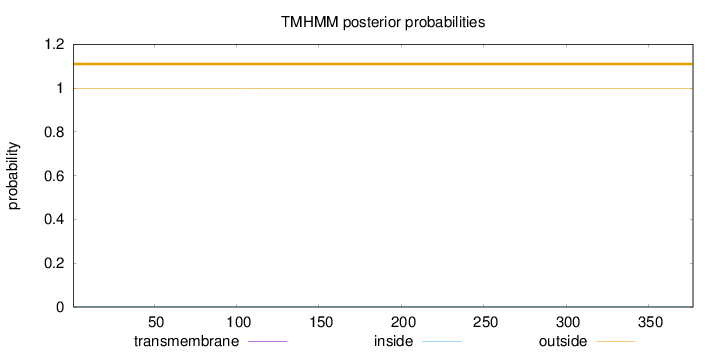
Topology
Subcellular location
Length:
377
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00093
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00108
outside
1 - 377
Population Genetic Test Statistics
Pi
17.75547
Theta
19.190003
Tajima's D
-0.428496
CLR
0.266904
CSRT
0.262786860656967
Interpretation
Uncertain
