Gene
KWMTBOMO12663
Annotation
PREDICTED:_ubiquitin-conjugating_enzyme_E2_J2-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.545 Extracellular Reliability : 1.373
Sequence
CDS
ATGGCAAAAACAAAGGTGACAGGAGCCACTAGCAGATTGAAGCAGGATTATTTGAGGCTCAAGAATGACCCAGTACCATATGTTACGGCCGAACCTGTGCCGTCCAACATACTGGAATGGCATTACGTCGTTAAAGGGCCGGAGAAAAGTCCCTACGAAGGGGGATATTATCATGGAAAGATTATTTTCCCAAGGGAATTTCCGTTCAAACCACCTTCGATTTACATGATAACACCGAATGGCAGGTTCAAGACAAACACACGATTGTGCCTCAGTATTACAGACTTTCATCCGGACACTTGGAACCCTGCTTGGTCTATTTCTACGATCCTTACAGGTCTGTTGAGTTTTATGTTGGAGAAGACTCCTACATTGGGGTCCATTGAAACTTCAGAATACCAAAAGCGATGTTTGGCTGCAGAATCTCTAGAGATAAACATAAAAAATAAAATGTTTTGTGAATTGTTCCCTGAATATGTAGAGGAGATTGAACAGCAATTAAAACAACGACAAGAGGCAATGTTGTTGAATCAAGACAGTGGTGCAAATGAAGTAACCTCTGAGCCACAGTCTAGTATGAATTGTATTTTGACAAATCTGTTTGTTCTTGTAGCTTTTGTATGCCTTACTTACATGGTCAAACATGTTTTAGGATCATATGAATAG
Protein
MAKTKVTGATSRLKQDYLRLKNDPVPYVTAEPVPSNILEWHYVVKGPEKSPYEGGYYHGKIIFPREFPFKPPSIYMITPNGRFKTNTRLCLSITDFHPDTWNPAWSISTILTGLLSFMLEKTPTLGSIETSEYQKRCLAAESLEINIKNKMFCELFPEYVEEIEQQLKQRQEAMLLNQDSGANEVTSEPQSSMNCILTNLFVLVAFVCLTYMVKHVLGSYE
Summary
Uniprot
A0A437BUX2
A0A2A4IUK0
A0A194RDX0
A0A212FFR1
A0A2H1VRZ5
A0A1E1WKW5
+ More
A0A0L7L475 A0A194QCH7 A0A1W4XIV8 E2C2E8 A0A154NY32 A0A195BBH2 A0A158NAC2 A0A195FUM5 F4WJU7 A0A026WS21 A0A151X4K6 E2AF39 A0A2A3EQF4 V9IKK9 A0A087ZP05 A0A0M8ZY93 A0A0L7R0I7 R7V2C0 A0A2I4AQ33 A0A1A8C6Q5 A0A1A8GQ49 A0A1A8UXE2 A0A1A7X1C7 A0A2U9BGH4 A0A3Q2ZDJ8 D6WL86 A0A1A8IXC1 A0A3Q4HBY8 A0A3Q2QCV1 I3KTC9 A0A1A8RSA6 A0A1A8QZ37 A0A3B4FDC8 A0A3Q2W8X7 A0A3P8P6W1 A0A3P9DPG5 T1G6I2 A0A2J7QC68 H2TQI9 A0A3Q0S9G5 T1DKC1 A0A1Y1NAT8 A0A3Q1JP65 J3S5E7 A0A067R6N3 G3NYP6 A4QNV0 A0A146ZZY4 A0A1S3EYW0 A0A3Q3KDS2 E6ZFF4 A0A3B3DSS1 A0A3Q2CXW9 A0A2F0BAN9 A0A383YUA5 A0A2Y9FRG8 A0A341CN21 A0A340WMT5 A0A2Y9QFP4 A0A250YDR8 A0A1U7R1S1 A0A1U7SRT9 W5M2U3 F7A451 A0A3Q3MPP6 J3JVN7 M7ASW0 G1M4W6 A0A3Q7Y9G0 H9GNU4 A0A1B6C965 G3I416 K9IHA1 A0A0P7UX40 A0A3B3S3P3 A0A0P6JAD1 A0A1U7RF38 A0A3B5PVL1 A0A3B5MHH0 A0A0G2K8C5 A0A1S2ZWQ9 L8J1R3 A0A452EJX9 F6TU09 A0A1W7RBZ6 A0A0S7GCT4 B1ASK8 A0A2Y9KA75 A0A3Q7U6A6 A0A0F8AK68 W5P6H1 A0A091MYV6 M3YRS2 A0A060YEI6
A0A0L7L475 A0A194QCH7 A0A1W4XIV8 E2C2E8 A0A154NY32 A0A195BBH2 A0A158NAC2 A0A195FUM5 F4WJU7 A0A026WS21 A0A151X4K6 E2AF39 A0A2A3EQF4 V9IKK9 A0A087ZP05 A0A0M8ZY93 A0A0L7R0I7 R7V2C0 A0A2I4AQ33 A0A1A8C6Q5 A0A1A8GQ49 A0A1A8UXE2 A0A1A7X1C7 A0A2U9BGH4 A0A3Q2ZDJ8 D6WL86 A0A1A8IXC1 A0A3Q4HBY8 A0A3Q2QCV1 I3KTC9 A0A1A8RSA6 A0A1A8QZ37 A0A3B4FDC8 A0A3Q2W8X7 A0A3P8P6W1 A0A3P9DPG5 T1G6I2 A0A2J7QC68 H2TQI9 A0A3Q0S9G5 T1DKC1 A0A1Y1NAT8 A0A3Q1JP65 J3S5E7 A0A067R6N3 G3NYP6 A4QNV0 A0A146ZZY4 A0A1S3EYW0 A0A3Q3KDS2 E6ZFF4 A0A3B3DSS1 A0A3Q2CXW9 A0A2F0BAN9 A0A383YUA5 A0A2Y9FRG8 A0A341CN21 A0A340WMT5 A0A2Y9QFP4 A0A250YDR8 A0A1U7R1S1 A0A1U7SRT9 W5M2U3 F7A451 A0A3Q3MPP6 J3JVN7 M7ASW0 G1M4W6 A0A3Q7Y9G0 H9GNU4 A0A1B6C965 G3I416 K9IHA1 A0A0P7UX40 A0A3B3S3P3 A0A0P6JAD1 A0A1U7RF38 A0A3B5PVL1 A0A3B5MHH0 A0A0G2K8C5 A0A1S2ZWQ9 L8J1R3 A0A452EJX9 F6TU09 A0A1W7RBZ6 A0A0S7GCT4 B1ASK8 A0A2Y9KA75 A0A3Q7U6A6 A0A0F8AK68 W5P6H1 A0A091MYV6 M3YRS2 A0A060YEI6
Pubmed
26354079
22118469
26227816
20798317
21347285
21719571
+ More
24508170 30249741 23254933 18362917 19820115 25186727 21551351 23758969 28004739 23025625 24845553 23594743 29451363 28087693 17495919 22516182 23537049 23624526 20010809 21881562 21804562 29240929 23542700 15057822 15632090 22751099 19892987 26358130 12040188 19468303 21183079 25835551 20809919 24755649
24508170 30249741 23254933 18362917 19820115 25186727 21551351 23758969 28004739 23025625 24845553 23594743 29451363 28087693 17495919 22516182 23537049 23624526 20010809 21881562 21804562 29240929 23542700 15057822 15632090 22751099 19892987 26358130 12040188 19468303 21183079 25835551 20809919 24755649
EMBL
RSAL01000006
RVE54267.1
NWSH01006503
PCG63391.1
KQ460325
KPJ15797.1
+ More
AGBW02008777 OWR52568.1 ODYU01003897 SOQ43232.1 GDQN01003439 GDQN01001317 JAT87615.1 JAT89737.1 JTDY01003039 KOB70272.1 KQ459193 KPJ03124.1 GL452124 EFN77898.1 KQ434781 KZC04491.1 KQ976528 KYM81883.1 ADTU01001464 KQ981264 KYN43992.1 GL888186 EGI65589.1 KK107119 QOIP01000009 EZA58743.1 RLU18829.1 KQ982544 KYQ55346.1 GL438984 EFN67994.1 KZ288204 PBC33271.1 JR051790 AEY61560.1 KQ435798 KOX73377.1 KQ414670 KOC64360.1 AMQN01005351 KB295796 ELU12627.1 HADZ01010812 HAEA01009619 SBP74753.1 HAEB01019236 HAEC01005051 SBQ73128.1 HADY01021169 HAEJ01012612 SBS53069.1 HADW01010330 HADX01000039 SBP11730.1 CP026248 AWP03113.1 KQ971343 EFA03490.1 HAED01015537 HAEE01013784 SBR01982.1 AERX01006852 HAEI01008080 HAEH01018711 SBS08114.1 HAEF01000210 HAEG01014890 SBR98777.1 AMQM01006822 KB097528 ESN95317.1 NEVH01016291 PNF26179.1 GAAZ01002792 JAA95151.1 GEZM01007641 GEZM01007640 JAV95024.1 JU176384 GBEX01003798 AFJ51907.1 JAI10762.1 KK852665 KDR18969.1 CR926464 BC139531 AAI39532.1 GCES01014432 JAR71891.1 FQ310506 CBN80904.1 NTJE010119119 MBW00051.1 GFFW01003025 JAV41763.1 AHAT01034620 AHAT01034621 AHAT01034622 APGK01021343 BT127305 KB740314 KB631743 AEE62267.1 ENN80907.1 ERL85714.1 KB585326 EMP25990.1 ACTA01044646 AAWZ02035685 GEDC01027324 JAS09974.1 JH001219 EGW07777.1 GABZ01007505 JAA46020.1 JARO02001701 KPP74669.1 GEBF01002989 JAO00644.1 AC126156 CH473968 EDL81345.1 JH880456 ELR61367.1 LWLT01000016 GDAY02002800 JAV48667.1 GBYX01455557 JAO25979.1 AL627204 CH466594 EDL15061.1 KQ041937 KKF20793.1 AMGL01021967 KK838948 KFP81852.1 AEYP01101970 FR909979 CDQ89932.1
AGBW02008777 OWR52568.1 ODYU01003897 SOQ43232.1 GDQN01003439 GDQN01001317 JAT87615.1 JAT89737.1 JTDY01003039 KOB70272.1 KQ459193 KPJ03124.1 GL452124 EFN77898.1 KQ434781 KZC04491.1 KQ976528 KYM81883.1 ADTU01001464 KQ981264 KYN43992.1 GL888186 EGI65589.1 KK107119 QOIP01000009 EZA58743.1 RLU18829.1 KQ982544 KYQ55346.1 GL438984 EFN67994.1 KZ288204 PBC33271.1 JR051790 AEY61560.1 KQ435798 KOX73377.1 KQ414670 KOC64360.1 AMQN01005351 KB295796 ELU12627.1 HADZ01010812 HAEA01009619 SBP74753.1 HAEB01019236 HAEC01005051 SBQ73128.1 HADY01021169 HAEJ01012612 SBS53069.1 HADW01010330 HADX01000039 SBP11730.1 CP026248 AWP03113.1 KQ971343 EFA03490.1 HAED01015537 HAEE01013784 SBR01982.1 AERX01006852 HAEI01008080 HAEH01018711 SBS08114.1 HAEF01000210 HAEG01014890 SBR98777.1 AMQM01006822 KB097528 ESN95317.1 NEVH01016291 PNF26179.1 GAAZ01002792 JAA95151.1 GEZM01007641 GEZM01007640 JAV95024.1 JU176384 GBEX01003798 AFJ51907.1 JAI10762.1 KK852665 KDR18969.1 CR926464 BC139531 AAI39532.1 GCES01014432 JAR71891.1 FQ310506 CBN80904.1 NTJE010119119 MBW00051.1 GFFW01003025 JAV41763.1 AHAT01034620 AHAT01034621 AHAT01034622 APGK01021343 BT127305 KB740314 KB631743 AEE62267.1 ENN80907.1 ERL85714.1 KB585326 EMP25990.1 ACTA01044646 AAWZ02035685 GEDC01027324 JAS09974.1 JH001219 EGW07777.1 GABZ01007505 JAA46020.1 JARO02001701 KPP74669.1 GEBF01002989 JAO00644.1 AC126156 CH473968 EDL81345.1 JH880456 ELR61367.1 LWLT01000016 GDAY02002800 JAV48667.1 GBYX01455557 JAO25979.1 AL627204 CH466594 EDL15061.1 KQ041937 KKF20793.1 AMGL01021967 KK838948 KFP81852.1 AEYP01101970 FR909979 CDQ89932.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000007151
UP000037510
UP000053268
+ More
UP000192223 UP000008237 UP000076502 UP000078540 UP000005205 UP000078541 UP000007755 UP000053097 UP000279307 UP000075809 UP000000311 UP000242457 UP000005203 UP000053105 UP000053825 UP000014760 UP000192220 UP000246464 UP000264800 UP000007266 UP000261580 UP000265000 UP000005207 UP000261460 UP000264840 UP000265100 UP000265160 UP000015101 UP000235965 UP000005226 UP000261340 UP000265040 UP000027135 UP000007635 UP000000437 UP000081671 UP000261600 UP000261560 UP000265020 UP000261681 UP000248484 UP000252040 UP000265300 UP000248483 UP000189706 UP000189704 UP000018468 UP000002280 UP000261640 UP000019118 UP000030742 UP000031443 UP000008912 UP000286642 UP000001646 UP000001075 UP000034805 UP000192224 UP000261540 UP000189705 UP000002852 UP000261380 UP000002494 UP000079721 UP000291000 UP000002281 UP000000589 UP000248482 UP000286640 UP000002356 UP000000715 UP000193380
UP000192223 UP000008237 UP000076502 UP000078540 UP000005205 UP000078541 UP000007755 UP000053097 UP000279307 UP000075809 UP000000311 UP000242457 UP000005203 UP000053105 UP000053825 UP000014760 UP000192220 UP000246464 UP000264800 UP000007266 UP000261580 UP000265000 UP000005207 UP000261460 UP000264840 UP000265100 UP000265160 UP000015101 UP000235965 UP000005226 UP000261340 UP000265040 UP000027135 UP000007635 UP000000437 UP000081671 UP000261600 UP000261560 UP000265020 UP000261681 UP000248484 UP000252040 UP000265300 UP000248483 UP000189706 UP000189704 UP000018468 UP000002280 UP000261640 UP000019118 UP000030742 UP000031443 UP000008912 UP000286642 UP000001646 UP000001075 UP000034805 UP000192224 UP000261540 UP000189705 UP000002852 UP000261380 UP000002494 UP000079721 UP000291000 UP000002281 UP000000589 UP000248482 UP000286640 UP000002356 UP000000715 UP000193380
PRIDE
Pfam
PF00179 UQ_con
SUPFAM
SSF54495
SSF54495
Gene 3D
CDD
ProteinModelPortal
A0A437BUX2
A0A2A4IUK0
A0A194RDX0
A0A212FFR1
A0A2H1VRZ5
A0A1E1WKW5
+ More
A0A0L7L475 A0A194QCH7 A0A1W4XIV8 E2C2E8 A0A154NY32 A0A195BBH2 A0A158NAC2 A0A195FUM5 F4WJU7 A0A026WS21 A0A151X4K6 E2AF39 A0A2A3EQF4 V9IKK9 A0A087ZP05 A0A0M8ZY93 A0A0L7R0I7 R7V2C0 A0A2I4AQ33 A0A1A8C6Q5 A0A1A8GQ49 A0A1A8UXE2 A0A1A7X1C7 A0A2U9BGH4 A0A3Q2ZDJ8 D6WL86 A0A1A8IXC1 A0A3Q4HBY8 A0A3Q2QCV1 I3KTC9 A0A1A8RSA6 A0A1A8QZ37 A0A3B4FDC8 A0A3Q2W8X7 A0A3P8P6W1 A0A3P9DPG5 T1G6I2 A0A2J7QC68 H2TQI9 A0A3Q0S9G5 T1DKC1 A0A1Y1NAT8 A0A3Q1JP65 J3S5E7 A0A067R6N3 G3NYP6 A4QNV0 A0A146ZZY4 A0A1S3EYW0 A0A3Q3KDS2 E6ZFF4 A0A3B3DSS1 A0A3Q2CXW9 A0A2F0BAN9 A0A383YUA5 A0A2Y9FRG8 A0A341CN21 A0A340WMT5 A0A2Y9QFP4 A0A250YDR8 A0A1U7R1S1 A0A1U7SRT9 W5M2U3 F7A451 A0A3Q3MPP6 J3JVN7 M7ASW0 G1M4W6 A0A3Q7Y9G0 H9GNU4 A0A1B6C965 G3I416 K9IHA1 A0A0P7UX40 A0A3B3S3P3 A0A0P6JAD1 A0A1U7RF38 A0A3B5PVL1 A0A3B5MHH0 A0A0G2K8C5 A0A1S2ZWQ9 L8J1R3 A0A452EJX9 F6TU09 A0A1W7RBZ6 A0A0S7GCT4 B1ASK8 A0A2Y9KA75 A0A3Q7U6A6 A0A0F8AK68 W5P6H1 A0A091MYV6 M3YRS2 A0A060YEI6
A0A0L7L475 A0A194QCH7 A0A1W4XIV8 E2C2E8 A0A154NY32 A0A195BBH2 A0A158NAC2 A0A195FUM5 F4WJU7 A0A026WS21 A0A151X4K6 E2AF39 A0A2A3EQF4 V9IKK9 A0A087ZP05 A0A0M8ZY93 A0A0L7R0I7 R7V2C0 A0A2I4AQ33 A0A1A8C6Q5 A0A1A8GQ49 A0A1A8UXE2 A0A1A7X1C7 A0A2U9BGH4 A0A3Q2ZDJ8 D6WL86 A0A1A8IXC1 A0A3Q4HBY8 A0A3Q2QCV1 I3KTC9 A0A1A8RSA6 A0A1A8QZ37 A0A3B4FDC8 A0A3Q2W8X7 A0A3P8P6W1 A0A3P9DPG5 T1G6I2 A0A2J7QC68 H2TQI9 A0A3Q0S9G5 T1DKC1 A0A1Y1NAT8 A0A3Q1JP65 J3S5E7 A0A067R6N3 G3NYP6 A4QNV0 A0A146ZZY4 A0A1S3EYW0 A0A3Q3KDS2 E6ZFF4 A0A3B3DSS1 A0A3Q2CXW9 A0A2F0BAN9 A0A383YUA5 A0A2Y9FRG8 A0A341CN21 A0A340WMT5 A0A2Y9QFP4 A0A250YDR8 A0A1U7R1S1 A0A1U7SRT9 W5M2U3 F7A451 A0A3Q3MPP6 J3JVN7 M7ASW0 G1M4W6 A0A3Q7Y9G0 H9GNU4 A0A1B6C965 G3I416 K9IHA1 A0A0P7UX40 A0A3B3S3P3 A0A0P6JAD1 A0A1U7RF38 A0A3B5PVL1 A0A3B5MHH0 A0A0G2K8C5 A0A1S2ZWQ9 L8J1R3 A0A452EJX9 F6TU09 A0A1W7RBZ6 A0A0S7GCT4 B1ASK8 A0A2Y9KA75 A0A3Q7U6A6 A0A0F8AK68 W5P6H1 A0A091MYV6 M3YRS2 A0A060YEI6
PDB
2F4W
E-value=2.09885e-76,
Score=724
Ontologies
PATHWAY
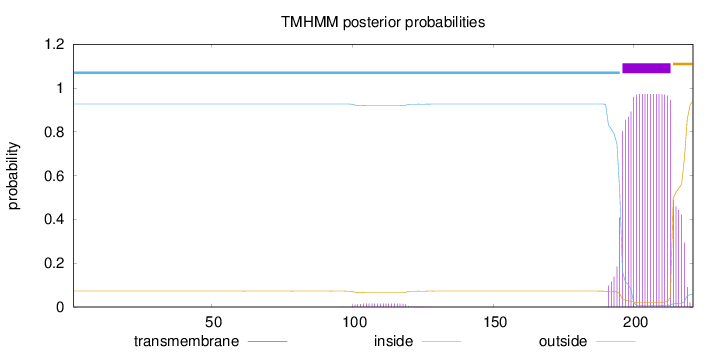
Topology
Length:
221
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.45101
Exp number, first 60 AAs:
0.00057
Total prob of N-in:
0.92725
inside
1 - 195
TMhelix
196 - 213
outside
214 - 221
Population Genetic Test Statistics
Pi
12.500897
Theta
11.487387
Tajima's D
-0.508295
CLR
0
CSRT
0.237538123093845
Interpretation
Uncertain
