Gene
KWMTBOMO12659 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001406
Annotation
PREDICTED:_T-complex_protein_1_subunit_gamma_[Amyelois_transitella]
Full name
T-complex protein 1 subunit gamma
Alternative Name
CCT-gamma
Chaperonin containing TCP1 subunit 3
Chaperonin containing TCP1 subunit 3
Location in the cell
Cytoplasmic Reliability : 3.545
Sequence
CDS
ATGTATGGACAGCAACCTATATTAGTTCTCAGCCAAAATACAAAACGTGAGTCCGGCCGGAAAGTTCAGCTTGAAAATATTAGTGCAGGAAAGACCATAGCCGATGTTATAAGAACATGCCTCGGACCTCAGGCCATGTTAAAAATGTTAATGGACCCTATGGGAGGCATCGTGATGACCAACGATGGTAATGCTATACTCAGGGAAATCACAGTCCAACATCCAGCTGCAAAATCTATGATCGAAATCGCAAGGACCCAAGATGAAGAGGTTGGAGATGGAACCACATCAGTCATAGTGCTAGCAGGAGAAATGTTGGCGATTGCAGAACCTTTCTTGACACAGAATATTCATCCAACAGTCATCATCAGAGAATACCGCCAAGCTTTAGAGGATGCTATAGTTCTGCTTCAAGACAAAATTTCAGTGCCTGTAGATTTGAATGATAGAGATAAGATGAAAGAAGTTATTCGTTCATGTGTTGGAACAAAGTACATCGGTCGTTGGGCAGATCTTGCCGTAGACATTGCTCTTGATGCGGTTAATACTGTTACTGTGAATGATAATGGCAGAATTGAGGTTGACATTAAAAATTATGCAAAAGTGGAAAAGATCCCAGGTGGCACAGTTGAAGAGTCTCGTGTACTCAGCGGTGTTATGTTCAACAAGGATGTAACCCATCCAAAGATGAGGAGGTATATTGAAAATCCGCGCATTATCCTGTTGGATTGCCCACTTGAATACAAAAAGGGAGAGAGTCAGACCAACATTGAGATTATGGGTGAACAGGACTTCACTAAACTGCTACAACTAGAAGAGGAACATGTACAGCGTCAATGTGAAGAAATTATTGCACTTAAACCAGATATTGTCTTTACCGAAAAGGGAGTTTCAGATCTTGCACAACATTACCTTGTTAAGGCTGGAATTACTGCTATCCGCAGACTCCGTAAAACGGACAACAACCGTCTAGCGCGGGCGTGTGGAGCAACAATTGTCAACCGTACTGAGGAACTCAAGGAATCCGATGTCGGCACTGGAGCTGGACTGTTTGAGATCAAGAAGATCGGAGATGAATATTTCACTTTTGTCACTGAATGCAAGAACCCCAAGGCATGCACAATATTACTCCGTGGTGCTTCTAAGGATGTCCTGAATGAGATTGAAAGGAATCTTCAAGACGCTTTACACGTCGCCAAGAATTTGGTGGTGTCGCCGCGGCTGGTGGCGGGGGGCGGCGCCGCGGAGATGGCCGTGTCCAGCGCGCTGTCGCACCGCGCCGCGCACAACACGCCCTACAGGGCCGTCGCCACCGCGCTCGAGATAATACCGCGCACGCTGGCGCAGAACTGCGGCGCCAACACCATCCGCACGCTCACGGCGCTGCGGGCGCGCCACGCCGCCGGACTCGCCTCCGCCGGCATCGACGGCGAGACCGGCGAGATCGTCGACATGGCCGTCAAGGGGATCTGGGAGCCGCTCGCCGTTAAACTGCAGGTGTACAAAACAGCCGTCGAAACAGCGATCCTGCTGTTGAGGATCGACGACATCGTGTCGGGCTCCAAGAAGAACAAGAACAACGAGCCCGCGGCGCCGGCCGAGATGGCCGCACAAGAAAACTAA
Protein
MYGQQPILVLSQNTKRESGRKVQLENISAGKTIADVIRTCLGPQAMLKMLMDPMGGIVMTNDGNAILREITVQHPAAKSMIEIARTQDEEVGDGTTSVIVLAGEMLAIAEPFLTQNIHPTVIIREYRQALEDAIVLLQDKISVPVDLNDRDKMKEVIRSCVGTKYIGRWADLAVDIALDAVNTVTVNDNGRIEVDIKNYAKVEKIPGGTVEESRVLSGVMFNKDVTHPKMRRYIENPRIILLDCPLEYKKGESQTNIEIMGEQDFTKLLQLEEEHVQRQCEEIIALKPDIVFTEKGVSDLAQHYLVKAGITAIRRLRKTDNNRLARACGATIVNRTEELKESDVGTGAGLFEIKKIGDEYFTFVTECKNPKACTILLRGASKDVLNEIERNLQDALHVAKNLVVSPRLVAGGGAAEMAVSSALSHRAAHNTPYRAVATALEIIPRTLAQNCGANTIRTLTALRARHAAGLASAGIDGETGEIVDMAVKGIWEPLAVKLQVYKTAVETAILLLRIDDIVSGSKKNKNNEPAAPAEMAAQEN
Summary
Subunit
Heterooligomeric complex of about 850 to 900 kDa that forms two stacked rings, 12 to 16 nm in diameter.
Similarity
Belongs to the TCP-1 chaperonin family.
Keywords
ATP-binding
Chaperone
Complete proteome
Cytoplasm
Disulfide bond
Nucleotide-binding
Reference proteome
Feature
chain T-complex protein 1 subunit gamma
Uniprot
H9IVX5
A0A2A4K5S4
A0A2H1VVJ7
A0A194QCD3
A0A437BUX5
S4P6K3
+ More
A0A2A4K7F4 W8B4Z1 A0A0A1WIQ5 A0A1Q3F384 A0A0K8UAV7 A0A0K8UQQ5 A0A034WB81 D1MDB9 A0A1L8ED77 B4LYG9 A0A1I8PYY9 A0A0L0BZX0 B4JG86 B0WSS5 A0A2M4A9B6 A0A2M4BJ12 A0A2M4BIY0 T1DSF4 B3M2K6 W5JIM5 A0A2M4BJ89 B4NJH6 A0A2M3Z6J9 A0A182G413 B4QWG0 A0A182P508 B4IBR9 Q16U15 A0A182M7U0 B4KAH9 Q297K0 A0A182VZQ2 A0A182F7D7 A0A3B0KAP5 A0A1W4UIV4 A0A182JW08 B3NZC4 A4V303 B4PMX4 P48605 A0A182RSY6 H6WAS3 A0A182NQG8 A0A182V0W7 A0A182LBZ2 A0A182XIU3 Q7QCQ5 A0A182HTH8 U5EM51 T1PFL6 A0A2P8YVF8 A0A0M5JC34 A0A182TLD8 D3TN28 A0A1A9WLU5 A0A1A9Z9W2 A0A1B0AS37 A0A182QK37 A0A182S668 A0A336MCC4 A0A1A9YHY4 A0A1A9VPD4 A0A2J7QC12 A0A084VTB1 A0A067RMU1 D6WRF8 A0A182J815 J3JVZ0 A0A1L8DUF6 V5RDL0 A0A0K8TS67 V5GRT0 A0A026W022 A0A1B6DCI8 A0A1B0GLJ4 E9IDA8 F4WXK8 A0A0J7KP09 A0A0L7R5R3 A0A151WTF3 N6TQ76 A0A195BVI5 E2BLS8 A0A158NUY2 A0A195D9R4 A0A1Y1JY21 A0A088A3C0 A0A1L8DUE6 A0A0C9Q0G4 A0A195FRP0 T1JFU3 E2A4G2 A0A154PBM0 A0A411G6C6
A0A2A4K7F4 W8B4Z1 A0A0A1WIQ5 A0A1Q3F384 A0A0K8UAV7 A0A0K8UQQ5 A0A034WB81 D1MDB9 A0A1L8ED77 B4LYG9 A0A1I8PYY9 A0A0L0BZX0 B4JG86 B0WSS5 A0A2M4A9B6 A0A2M4BJ12 A0A2M4BIY0 T1DSF4 B3M2K6 W5JIM5 A0A2M4BJ89 B4NJH6 A0A2M3Z6J9 A0A182G413 B4QWG0 A0A182P508 B4IBR9 Q16U15 A0A182M7U0 B4KAH9 Q297K0 A0A182VZQ2 A0A182F7D7 A0A3B0KAP5 A0A1W4UIV4 A0A182JW08 B3NZC4 A4V303 B4PMX4 P48605 A0A182RSY6 H6WAS3 A0A182NQG8 A0A182V0W7 A0A182LBZ2 A0A182XIU3 Q7QCQ5 A0A182HTH8 U5EM51 T1PFL6 A0A2P8YVF8 A0A0M5JC34 A0A182TLD8 D3TN28 A0A1A9WLU5 A0A1A9Z9W2 A0A1B0AS37 A0A182QK37 A0A182S668 A0A336MCC4 A0A1A9YHY4 A0A1A9VPD4 A0A2J7QC12 A0A084VTB1 A0A067RMU1 D6WRF8 A0A182J815 J3JVZ0 A0A1L8DUF6 V5RDL0 A0A0K8TS67 V5GRT0 A0A026W022 A0A1B6DCI8 A0A1B0GLJ4 E9IDA8 F4WXK8 A0A0J7KP09 A0A0L7R5R3 A0A151WTF3 N6TQ76 A0A195BVI5 E2BLS8 A0A158NUY2 A0A195D9R4 A0A1Y1JY21 A0A088A3C0 A0A1L8DUE6 A0A0C9Q0G4 A0A195FRP0 T1JFU3 E2A4G2 A0A154PBM0 A0A411G6C6
Pubmed
19121390
26354079
23622113
24495485
25830018
25348373
+ More
19936241 17994087 26108605 20920257 23761445 18057021 26483478 17510324 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 7667301 8666276 12537569 23055932 25244985 20966253 12364791 14747013 17210077 25315136 29403074 20353571 24438588 24845553 18362917 19820115 22516182 26369729 24508170 30249741 21282665 21719571 23537049 20798317 21347285 28004739
19936241 17994087 26108605 20920257 23761445 18057021 26483478 17510324 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 7667301 8666276 12537569 23055932 25244985 20966253 12364791 14747013 17210077 25315136 29403074 20353571 24438588 24845553 18362917 19820115 22516182 26369729 24508170 30249741 21282665 21719571 23537049 20798317 21347285 28004739
EMBL
BABH01006486
NWSH01000099
PCG79581.1
ODYU01004392
SOQ44224.1
KQ459193
+ More
KPJ03127.1 RSAL01000006 RVE54268.1 GAIX01004999 JAA87561.1 PCG79582.1 GAMC01014405 GAMC01014402 JAB92153.1 GBXI01015550 JAC98741.1 GFDL01013087 JAV21958.1 GDHF01028550 JAI23764.1 GDHF01023285 JAI29029.1 GAKP01007399 GAKP01007395 JAC51557.1 GU132453 ACZ50717.1 GFDG01002121 JAV16678.1 CH940650 EDW66965.1 JRES01001097 KNC25531.1 CH916369 EDV93653.1 DS232075 EDS34034.1 GGFK01004030 MBW37351.1 GGFJ01003881 MBW53022.1 GGFJ01003878 MBW53019.1 GAMD01001509 JAB00082.1 CH902617 EDV42327.1 ADMH02001371 ETN62745.1 GGFJ01003880 MBW53021.1 CH964272 EDW83900.1 KRF99916.1 GGFM01003380 MBW24131.1 JXUM01041709 KQ561294 KXJ79086.1 CM000364 EDX12651.1 CH480827 EDW44827.1 CH477635 EAT38003.1 AXCM01000485 CH933806 EDW15692.1 CM000070 EAL28205.1 OUUW01000007 SPP83169.1 CH954181 EDV48796.1 AE014297 AAN13716.1 CM000160 EDW96986.1 U31961 X95602 AY089543 JQ042688 AEZ67841.1 AAAB01008859 EAA07808.4 APCN01000532 GANO01001146 JAB58725.1 KA647597 AFP62226.1 PYGN01000338 PSN48230.1 CP012526 ALC45668.1 CCAG010004095 EZ422830 ADD19106.1 JXJN01002668 AXCN02001530 UFQT01000755 UFQT01000940 SSX26991.1 SSX28111.1 NEVH01016292 PNF26121.1 ATLV01016287 KE525074 KFB41205.1 KK852415 KDR24373.1 KQ971351 EFA06459.1 BT127408 AEE62370.1 GFDF01004020 JAV10064.1 KF621134 AHB33470.1 GDAI01000406 JAI17197.1 GALX01004124 JAB64342.1 KK107519 QOIP01000009 EZA49403.1 RLU19095.1 GEDC01013973 JAS23325.1 AJWK01035082 AJWK01035083 AJWK01035084 AJWK01035085 AJWK01035086 GL762454 EFZ21433.1 GL888425 EGI61078.1 LBMM01004803 KMQ92073.1 KQ414648 KOC66154.1 KQ982769 KYQ50915.1 APGK01025209 KB740577 KB631756 ENN80183.1 ERL85819.1 KQ976401 KYM92634.1 GL449036 EFN83412.1 ADTU01026823 KQ981082 KYN09611.1 GEZM01097423 JAV54214.1 GFDF01004025 JAV10059.1 GBYB01007323 JAG77090.1 KQ981335 KYN42579.1 KYN42593.1 JH432185 GL436635 EFN71709.1 KQ434869 KZC09241.1 MH365604 QBB01413.1
KPJ03127.1 RSAL01000006 RVE54268.1 GAIX01004999 JAA87561.1 PCG79582.1 GAMC01014405 GAMC01014402 JAB92153.1 GBXI01015550 JAC98741.1 GFDL01013087 JAV21958.1 GDHF01028550 JAI23764.1 GDHF01023285 JAI29029.1 GAKP01007399 GAKP01007395 JAC51557.1 GU132453 ACZ50717.1 GFDG01002121 JAV16678.1 CH940650 EDW66965.1 JRES01001097 KNC25531.1 CH916369 EDV93653.1 DS232075 EDS34034.1 GGFK01004030 MBW37351.1 GGFJ01003881 MBW53022.1 GGFJ01003878 MBW53019.1 GAMD01001509 JAB00082.1 CH902617 EDV42327.1 ADMH02001371 ETN62745.1 GGFJ01003880 MBW53021.1 CH964272 EDW83900.1 KRF99916.1 GGFM01003380 MBW24131.1 JXUM01041709 KQ561294 KXJ79086.1 CM000364 EDX12651.1 CH480827 EDW44827.1 CH477635 EAT38003.1 AXCM01000485 CH933806 EDW15692.1 CM000070 EAL28205.1 OUUW01000007 SPP83169.1 CH954181 EDV48796.1 AE014297 AAN13716.1 CM000160 EDW96986.1 U31961 X95602 AY089543 JQ042688 AEZ67841.1 AAAB01008859 EAA07808.4 APCN01000532 GANO01001146 JAB58725.1 KA647597 AFP62226.1 PYGN01000338 PSN48230.1 CP012526 ALC45668.1 CCAG010004095 EZ422830 ADD19106.1 JXJN01002668 AXCN02001530 UFQT01000755 UFQT01000940 SSX26991.1 SSX28111.1 NEVH01016292 PNF26121.1 ATLV01016287 KE525074 KFB41205.1 KK852415 KDR24373.1 KQ971351 EFA06459.1 BT127408 AEE62370.1 GFDF01004020 JAV10064.1 KF621134 AHB33470.1 GDAI01000406 JAI17197.1 GALX01004124 JAB64342.1 KK107519 QOIP01000009 EZA49403.1 RLU19095.1 GEDC01013973 JAS23325.1 AJWK01035082 AJWK01035083 AJWK01035084 AJWK01035085 AJWK01035086 GL762454 EFZ21433.1 GL888425 EGI61078.1 LBMM01004803 KMQ92073.1 KQ414648 KOC66154.1 KQ982769 KYQ50915.1 APGK01025209 KB740577 KB631756 ENN80183.1 ERL85819.1 KQ976401 KYM92634.1 GL449036 EFN83412.1 ADTU01026823 KQ981082 KYN09611.1 GEZM01097423 JAV54214.1 GFDF01004025 JAV10059.1 GBYB01007323 JAG77090.1 KQ981335 KYN42579.1 KYN42593.1 JH432185 GL436635 EFN71709.1 KQ434869 KZC09241.1 MH365604 QBB01413.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000008792
UP000095300
+ More
UP000037069 UP000001070 UP000002320 UP000007801 UP000000673 UP000007798 UP000069940 UP000249989 UP000000304 UP000075885 UP000001292 UP000008820 UP000075883 UP000009192 UP000001819 UP000075920 UP000069272 UP000268350 UP000192221 UP000075881 UP000008711 UP000000803 UP000002282 UP000075900 UP000076408 UP000075884 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000095301 UP000245037 UP000092553 UP000075902 UP000092444 UP000091820 UP000092445 UP000092460 UP000075886 UP000075901 UP000092443 UP000078200 UP000235965 UP000030765 UP000027135 UP000007266 UP000075880 UP000053097 UP000279307 UP000092461 UP000007755 UP000036403 UP000053825 UP000075809 UP000019118 UP000030742 UP000078540 UP000008237 UP000005205 UP000078492 UP000005203 UP000078541 UP000000311 UP000076502
UP000037069 UP000001070 UP000002320 UP000007801 UP000000673 UP000007798 UP000069940 UP000249989 UP000000304 UP000075885 UP000001292 UP000008820 UP000075883 UP000009192 UP000001819 UP000075920 UP000069272 UP000268350 UP000192221 UP000075881 UP000008711 UP000000803 UP000002282 UP000075900 UP000076408 UP000075884 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000095301 UP000245037 UP000092553 UP000075902 UP000092444 UP000091820 UP000092445 UP000092460 UP000075886 UP000075901 UP000092443 UP000078200 UP000235965 UP000030765 UP000027135 UP000007266 UP000075880 UP000053097 UP000279307 UP000092461 UP000007755 UP000036403 UP000053825 UP000075809 UP000019118 UP000030742 UP000078540 UP000008237 UP000005205 UP000078492 UP000005203 UP000078541 UP000000311 UP000076502
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IVX5
A0A2A4K5S4
A0A2H1VVJ7
A0A194QCD3
A0A437BUX5
S4P6K3
+ More
A0A2A4K7F4 W8B4Z1 A0A0A1WIQ5 A0A1Q3F384 A0A0K8UAV7 A0A0K8UQQ5 A0A034WB81 D1MDB9 A0A1L8ED77 B4LYG9 A0A1I8PYY9 A0A0L0BZX0 B4JG86 B0WSS5 A0A2M4A9B6 A0A2M4BJ12 A0A2M4BIY0 T1DSF4 B3M2K6 W5JIM5 A0A2M4BJ89 B4NJH6 A0A2M3Z6J9 A0A182G413 B4QWG0 A0A182P508 B4IBR9 Q16U15 A0A182M7U0 B4KAH9 Q297K0 A0A182VZQ2 A0A182F7D7 A0A3B0KAP5 A0A1W4UIV4 A0A182JW08 B3NZC4 A4V303 B4PMX4 P48605 A0A182RSY6 H6WAS3 A0A182NQG8 A0A182V0W7 A0A182LBZ2 A0A182XIU3 Q7QCQ5 A0A182HTH8 U5EM51 T1PFL6 A0A2P8YVF8 A0A0M5JC34 A0A182TLD8 D3TN28 A0A1A9WLU5 A0A1A9Z9W2 A0A1B0AS37 A0A182QK37 A0A182S668 A0A336MCC4 A0A1A9YHY4 A0A1A9VPD4 A0A2J7QC12 A0A084VTB1 A0A067RMU1 D6WRF8 A0A182J815 J3JVZ0 A0A1L8DUF6 V5RDL0 A0A0K8TS67 V5GRT0 A0A026W022 A0A1B6DCI8 A0A1B0GLJ4 E9IDA8 F4WXK8 A0A0J7KP09 A0A0L7R5R3 A0A151WTF3 N6TQ76 A0A195BVI5 E2BLS8 A0A158NUY2 A0A195D9R4 A0A1Y1JY21 A0A088A3C0 A0A1L8DUE6 A0A0C9Q0G4 A0A195FRP0 T1JFU3 E2A4G2 A0A154PBM0 A0A411G6C6
A0A2A4K7F4 W8B4Z1 A0A0A1WIQ5 A0A1Q3F384 A0A0K8UAV7 A0A0K8UQQ5 A0A034WB81 D1MDB9 A0A1L8ED77 B4LYG9 A0A1I8PYY9 A0A0L0BZX0 B4JG86 B0WSS5 A0A2M4A9B6 A0A2M4BJ12 A0A2M4BIY0 T1DSF4 B3M2K6 W5JIM5 A0A2M4BJ89 B4NJH6 A0A2M3Z6J9 A0A182G413 B4QWG0 A0A182P508 B4IBR9 Q16U15 A0A182M7U0 B4KAH9 Q297K0 A0A182VZQ2 A0A182F7D7 A0A3B0KAP5 A0A1W4UIV4 A0A182JW08 B3NZC4 A4V303 B4PMX4 P48605 A0A182RSY6 H6WAS3 A0A182NQG8 A0A182V0W7 A0A182LBZ2 A0A182XIU3 Q7QCQ5 A0A182HTH8 U5EM51 T1PFL6 A0A2P8YVF8 A0A0M5JC34 A0A182TLD8 D3TN28 A0A1A9WLU5 A0A1A9Z9W2 A0A1B0AS37 A0A182QK37 A0A182S668 A0A336MCC4 A0A1A9YHY4 A0A1A9VPD4 A0A2J7QC12 A0A084VTB1 A0A067RMU1 D6WRF8 A0A182J815 J3JVZ0 A0A1L8DUF6 V5RDL0 A0A0K8TS67 V5GRT0 A0A026W022 A0A1B6DCI8 A0A1B0GLJ4 E9IDA8 F4WXK8 A0A0J7KP09 A0A0L7R5R3 A0A151WTF3 N6TQ76 A0A195BVI5 E2BLS8 A0A158NUY2 A0A195D9R4 A0A1Y1JY21 A0A088A3C0 A0A1L8DUE6 A0A0C9Q0G4 A0A195FRP0 T1JFU3 E2A4G2 A0A154PBM0 A0A411G6C6
PDB
4B2T
E-value=0,
Score=1793
Ontologies
KEGG
GO
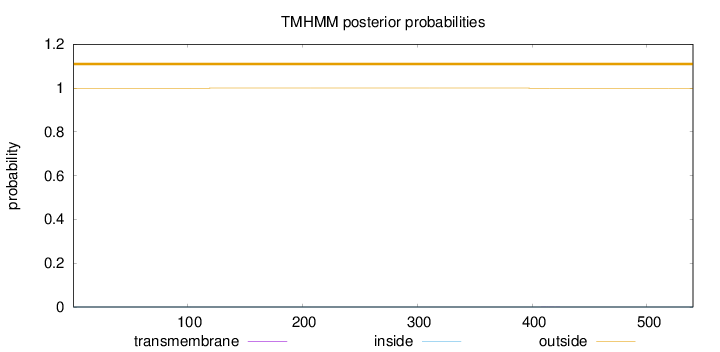
Topology
Subcellular location
Cytoplasm
Length:
540
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00664999999999999
Exp number, first 60 AAs:
0.00104
Total prob of N-in:
0.00050
outside
1 - 540
Population Genetic Test Statistics
Pi
28.21734
Theta
26.307183
Tajima's D
0.222832
CLR
0.913848
CSRT
0.44172791360432
Interpretation
Uncertain
